Project Description
It has been shown that mouse fibroblasts efficiently dedifferentiate and express markers of pluripotency after being geometrically confined over 10 days, without exogenous biochemical factors. After 6 days mesenchymal genes are repressed, and after 10 days ESC and iPSC specific genes are expressed. This work was published in PNAS in 2018.
More information on the Shivashankar Lab at MBI.
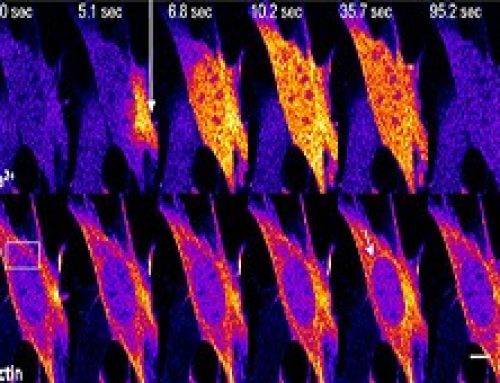
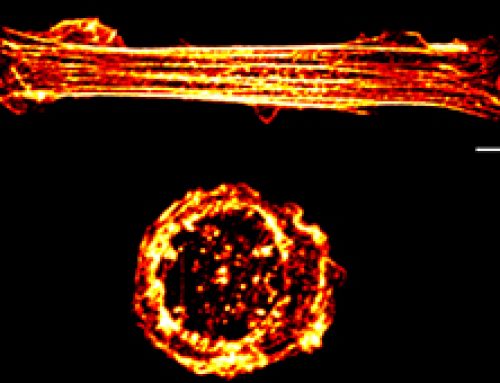
Figure: After growing mouse fibroblasts on confined geometries, spheroids developed. Over 10 days the cells began expressing ESC and iPSC genes, and reduced the expression of mesenchymal genes. These cells could then be directed down endoderm or neural ectoderm lineages.
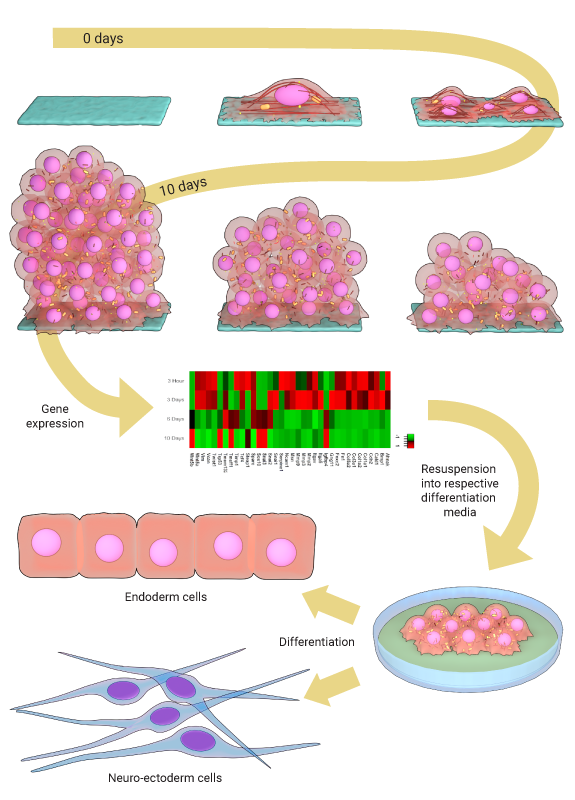
Mature fibroblast cells can be reprogrammed, in vitro, to acquire stemness or pluripotency by transfecting them with transcription factors such as the Oct-4, Sox2, c-Myc and Klf4. However, in vivo, mature cells may dedifferentiate during development, or to facilitate tissue maintenance, and this occurs in the absence of exogenous biochemical factors. Roy et al show that fibroblasts can be reprogrammed to acquire pluripotency by confining them to a defined geometry, which is achieved by culturing cells on ECM coated micropatterned substrates. The cells, left to develop into a spheroid over 10 days, exhibited an increase in nuclear plasticity and chromatin reorganization, with a reduction in the level of H3K9Ac at mesenchymal promoters and a subsequent reduction in the expression of mesenchymal genes at day 6. The expression of stem cell specific genes such as Oct 4 and Sox2 increased at day 6, before being maximally expressed at day 10. To show the differentiation potential of the spheroids, the cells were directed into either endoderm or neuro-ectoderm linages using the respective differentiation media.
Understanding the basics
Stem cells are unspecialized cells that have the potential to specialize or differentiate into different cell types in a multicellular organism. They are also defined by their ability to continuously replenish their populations by going through unlimited cycles of cell divisions. They occur as embryonic stem cells, which differentiate into the three germ layers that then evolve into specialized cell types forming the various tissues and organs in the body, and as adult stem cells, which serve as a source for replenishing old or damaged tissues during repair processes. Based on the number of cell lineages into which stem cells can differentiate into, they are classified as totipotent (differentiates into all cell types and the extra-embryonic cells), pluripotent (differentiate into all cell types), multipotent (differentiates into many cell cell lineages), oligopotent (differentiates into a few cell lineages), or unipotent (differentiates into a single cell lineage).
Pluripotent stem cells found during early organism development commonly exhibit softer nuclei, lack type-A lamins, and have a poorly defined cytoskeleton. Their chromatin is less compact and more transcription-permissive compared to differentiated cells. Although much is not known about the molecular signature of stem cells, Oct4, Sox2, and Nanog have been identified as some of the most commonly expressed embryonic stem cell genes. As the organism develops, pluripotent embryonic stem cells gradually undergo lineage restriction and transform into stem cells with limited differentiation capacities. This results in cell type-specific transcriptional profiles that is defined by the mechanical state of the nuclei as well as the epigenetic modifications on their chromatin structures.
Cells sense the physical properties of their substrates via cell-matrix adhesion complexes, especially integrin-based focal adhesions. Depending on external cues such as substrate stiffness and topology, different force-induced signaling pathways are mediated by focal adhesions that control fundamental cell functions, including its fate or lineage specification. Recent studies studying the role of substrate rigidity on cell fate have shown how cells cultured on substrates of different stiffness are reprogrammed into specific cell lineages. For instance, cells grown on softer substrates mimicking brain differentiate into a neuronal lineage, while cells grown on stiffer substrates mimicking muscles differentiate into a myogenic lineage and those grown on more rigid substrates mimicking bones specialize into bone cells.
The Study in Detail
Key Findings
- The most effective geometric constraint with which to induce nuclear reprogramming was a 1800um2 rectangle. This was compared to circular geometries of 1800um2 and 500um2. Spheroids grew on the rectangular pattern with an efficiency of 20%.
- The presence of H3K9Ac, a marker of transcription, was assessed at the promoter regions of 84 genes. It gradually reduced at some mesenchymal genes, including Snai2/3, Twist1 and Sox10, but was increased in genes associated with cell proliferation, epithelial genes, and reprogramming genes. Globally the average level of H3k9ac gradually increased from day 3 to day 10.
- After 6 days of growth, mesenchymal associated genes were repressed. A sharp increase in the expression of genes associated with embryonic stem cells and induced pluripotent stem cells was noted at Day 6, reaching a maximal 8-fold median increase after 10 days of growth.
- Changes in the gene expression profiles saw a decrease over the first 6 days in genes associated with cytoskeleton dynamics, including actin filament organization and contraction. Later, it was noted that the activity of the transcriptional repressor SMAD3 was reduced, while the transcription factors Oct-4 and SOX2, as well as several members of the WNT and BMP-SMAD pathway, were increased. It was proposed that these pathways, when activated by geometric confinement, are essential in the induction and maintenance of pluripotency.
- When resuspended together with the stem cell growth factor LIF, cells from the spheroids could be maintained for several days. Without LIF, and when plated on fibronectin, the cells exhibited a phenotype consistent with fibroblasts, with a higher expression (compared to control cells) of fibroblast associated genes Twist1, Acta2 and Snai1.
- When grown in the respective differentiation media, 10-day spheroid cells could be directed down a neural ectoderm or endoderm linage.
- To examine the effect of geometric confinement on cancer cells, geometrically confined MCF-7 cells were grown for 10 days and their gene expression profiles examined. It was found that confinement facilitated the induction of cancer stemness, with the gene expression profiles of 10 day old MCF7 spheroids mimicking that of a mammosphere.
Methods and Controls used in the study
- To enable geometric confinement of fibroblasts and MCF-7 cells, microcontact printing of fibronectin was used to create rectangular and circular patches.
- Stemness of spheroids was determined by staining for alkaline phosphatase activity.
- The occupancy of H39KAc at the promoters of 84 genes was assessed by an Epitect-ChIP qPCR assay using H3K9Ac antibodies and the mRNA levels of various transcription factors were assessed at various time points using qRT-PCR.
- Differentiation of resuspended spheroid cells was achieved using a commercially available differentiation media, with gene expression profiles compared against parental NIH3T3 cells.
Applications and Future Directions
The methods described in this study successfully induced pluripotency using geometrical confinement alone, and therefore without requiring cells to be transfected with transcription factors. It is hoped that the methods described in this study will lead to improved stem cell technologies for tissue engineering and regenerative medicine.