DNA packaging
What are nucleosomes?[Edit]
In order to fit DNA into the nucleus, it must be packaged into a highly compacted structure known as chromatin. In the first step of this process DNA is condensed into a 11 nm fiber that represents an approximate 6-fold level of compaction [1]. This is achieved through nucleosome assembly.
 Figure 1. Nucleosome is the first level of DNA packaging:: Each nucleosome consists of histone octamer core, assembled from the histones H2A, H2B, H3 and H4 (or other histone variants in some cases) and a segment of DNA that wraps around the histone core. Adjacent nucleosomes are connected via “linker DNA”.
The nucleosome is the smallest structural component of chromatin, and is produced through interactions between DNA and histone proteins. Here, a histone octamer is formed from the histones H2A, H2B, H3 and H4, although in some cases other histone variants may also be found in the core (e.g., H2A.Z, MacroH2A, H2a.Bbd, H2A.lap1, H2A.X, H3.3, CenH3 and others [1]). A 147bp segment of DNA then wraps around the histone octamer 1.75 times, thus completing the formation of a single nucleosome.
Figure 1. Nucleosome is the first level of DNA packaging:: Each nucleosome consists of histone octamer core, assembled from the histones H2A, H2B, H3 and H4 (or other histone variants in some cases) and a segment of DNA that wraps around the histone core. Adjacent nucleosomes are connected via “linker DNA”.
The nucleosome is the smallest structural component of chromatin, and is produced through interactions between DNA and histone proteins. Here, a histone octamer is formed from the histones H2A, H2B, H3 and H4, although in some cases other histone variants may also be found in the core (e.g., H2A.Z, MacroH2A, H2a.Bbd, H2A.lap1, H2A.X, H3.3, CenH3 and others [1]). A 147bp segment of DNA then wraps around the histone octamer 1.75 times, thus completing the formation of a single nucleosome.
Of course, a single nucleosome will not form in isolation but is instead part of a wider process, whereby multiple nucleosomes form in a linear fashion along the DNA molecule. This ultimately produces the 11 nm fiber, which is traditionally described, based on its appearance, as “beads on a string” [2]. Here, adjacent nucleosomes are connected via “linker DNA”, which is usually bound to the H1 histone and is between 20-80 bps long. Additionally, flexible histone tails which originate from the histone octamer extend away from nucleosomal DNA and can interact with other nucleosomes, stabilizing more complex 3D structures [3]. In other words, specific nucleosomes can be far apart with respect to their linear sequence, but within interacting distance in the context of higher order chromatin structure [1].
Alternative nucleosome conformations (reviewed in [1]) may arise due to spontaneous unwrapping and rewrapping of DNA around the histone core, as well as due to variations in histones themselves. Moreover, nucleosomes are highly dynamic and can undergo spontaneous sliding, “splitting” or even complete dissociation.
The level of compaction attained through the formation of the 11 nm nucleosome fiber is insufficient to package the whole genome into the nucleus. Instead, this fiber forms the basis for other higher order chromatin structures that are established through additional folding and bending events.
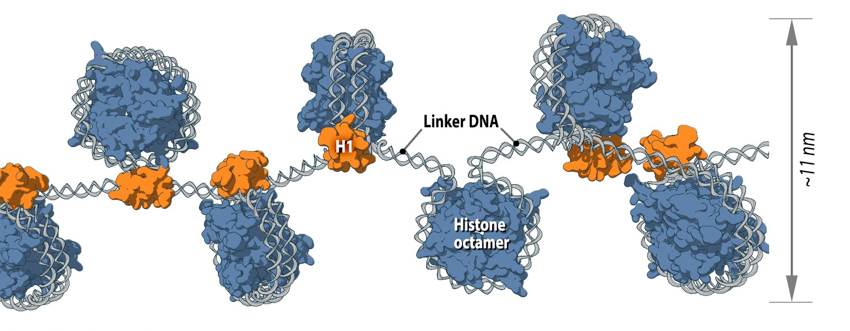 Figure 1. Nucleosome is the first level of DNA packaging:: Each nucleosome consists of histone octamer core, assembled from the histones H2A, H2B, H3 and H4 (or other histone variants in some cases) and a segment of DNA that wraps around the histone core. Adjacent nucleosomes are connected via “linker DNA”.
Figure 1. Nucleosome is the first level of DNA packaging:: Each nucleosome consists of histone octamer core, assembled from the histones H2A, H2B, H3 and H4 (or other histone variants in some cases) and a segment of DNA that wraps around the histone core. Adjacent nucleosomes are connected via “linker DNA”.Of course, a single nucleosome will not form in isolation but is instead part of a wider process, whereby multiple nucleosomes form in a linear fashion along the DNA molecule. This ultimately produces the 11 nm fiber, which is traditionally described, based on its appearance, as “beads on a string” [2]. Here, adjacent nucleosomes are connected via “linker DNA”, which is usually bound to the H1 histone and is between 20-80 bps long. Additionally, flexible histone tails which originate from the histone octamer extend away from nucleosomal DNA and can interact with other nucleosomes, stabilizing more complex 3D structures [3]. In other words, specific nucleosomes can be far apart with respect to their linear sequence, but within interacting distance in the context of higher order chromatin structure [1].
Alternative nucleosome conformations (reviewed in [1]) may arise due to spontaneous unwrapping and rewrapping of DNA around the histone core, as well as due to variations in histones themselves. Moreover, nucleosomes are highly dynamic and can undergo spontaneous sliding, “splitting” or even complete dissociation.
The level of compaction attained through the formation of the 11 nm nucleosome fiber is insufficient to package the whole genome into the nucleus. Instead, this fiber forms the basis for other higher order chromatin structures that are established through additional folding and bending events.
Intermediate chromatin structures[Edit]
Despite the extensive knowledge already gained on the structure of the 11 nm nucleosome fiber, as well as metaphase chromosomes, the intermediate chromatin structures commonly described are largely hypothetical and yet to be observed in vivo.
 Figure 2. Solenoid (A) and zigzag (B) models of intermediate chromatin condensation.: 30 nm chromatin fibers are considered to exist in the form of so called solenoid or zigzag. The main feature of solenoid model is that nucleosomes follow each other along the same helical path, and interactions between the histone cores occur sequentially (1, 2, 3 and so on). Therefore, solenoid is also referred to as “one start model”. In zigzag, on the other hand, linker DNA connects two opposing nucleosomes, creating a structure where the alternate histone cores become interacting partners (i.e., 1 and 3, 2 and 4 and so on). Therefore, zigzag is considered as a “two start model”, which is indicated in the figure (B) by two different colors of histone cores: yellow interacting nucleosome partners (1, 3, etc.) as opposed to the violet nucleosome row (2, 4, etc.).
Figure 2. Solenoid (A) and zigzag (B) models of intermediate chromatin condensation.: 30 nm chromatin fibers are considered to exist in the form of so called solenoid or zigzag. The main feature of solenoid model is that nucleosomes follow each other along the same helical path, and interactions between the histone cores occur sequentially (1, 2, 3 and so on). Therefore, solenoid is also referred to as “one start model”. In zigzag, on the other hand, linker DNA connects two opposing nucleosomes, creating a structure where the alternate histone cores become interacting partners (i.e., 1 and 3, 2 and 4 and so on). Therefore, zigzag is considered as a “two start model”, which is indicated in the figure (B) by two different colors of histone cores: yellow interacting nucleosome partners (1, 3, etc.) as opposed to the violet nucleosome row (2, 4, etc.).
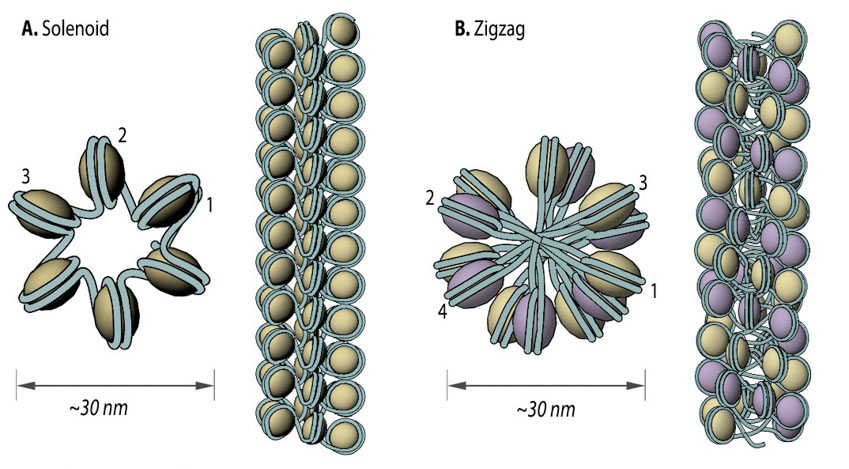 Figure 2. Solenoid (A) and zigzag (B) models of intermediate chromatin condensation.: 30 nm chromatin fibers are considered to exist in the form of so called solenoid or zigzag. The main feature of solenoid model is that nucleosomes follow each other along the same helical path, and interactions between the histone cores occur sequentially (1, 2, 3 and so on). Therefore, solenoid is also referred to as “one start model”. In zigzag, on the other hand, linker DNA connects two opposing nucleosomes, creating a structure where the alternate histone cores become interacting partners (i.e., 1 and 3, 2 and 4 and so on). Therefore, zigzag is considered as a “two start model”, which is indicated in the figure (B) by two different colors of histone cores: yellow interacting nucleosome partners (1, 3, etc.) as opposed to the violet nucleosome row (2, 4, etc.).
Figure 2. Solenoid (A) and zigzag (B) models of intermediate chromatin condensation.: 30 nm chromatin fibers are considered to exist in the form of so called solenoid or zigzag. The main feature of solenoid model is that nucleosomes follow each other along the same helical path, and interactions between the histone cores occur sequentially (1, 2, 3 and so on). Therefore, solenoid is also referred to as “one start model”. In zigzag, on the other hand, linker DNA connects two opposing nucleosomes, creating a structure where the alternate histone cores become interacting partners (i.e., 1 and 3, 2 and 4 and so on). Therefore, zigzag is considered as a “two start model”, which is indicated in the figure (B) by two different colors of histone cores: yellow interacting nucleosome partners (1, 3, etc.) as opposed to the violet nucleosome row (2, 4, etc.).Two popular models that were proposed based on in vitro data are the solenoid and zigzag. In each case, the 11 nm nucleosome fiber undergoes additional folding to form a 30 nm fiber [4, 5] with the manner of folding for a particular region depending on the internucleosomal linker length and the presence of linker histone H17 [6]. In the one-start solenoid model, bent linker DNA sequentially connects each nucleosome cores, creating a structure where nucleosomes follow each other along the same helical path [4, 7]. Alternatively, in the two-start zigzag model, straight linker DNA connects two opposing nucleosome cores, creating the opposing rows of nucleosomes that form so called “two-start” helix. In zigzag model, alternate nucleosomes (for example, N1 and N3) become interacting partners [5, 8]. Interestingly, some studies offer a model, where intermediate 30 nm fibers contain both the solenoid and zigzag conformations [9], suggesting instead that observations made in in vitro experiments might be an isolation artifact due to strictly cationic low-salt environment or chemical cross-linking (e.g., glutaraldehyde fixation). Consequently, new models of 11 nm fiber compaction have been proposed (e.g., chromonema, chromatin hub, hybrid chromonema/chromatin hub, fractal [10, 11, 12]), but no common conclusion has been reached yet.
One aspect shared by most of the models for higher order chromatin organization is the dynamic existence of decondensed loops among more compact chromatin structures. In most cases, higher order chromatin has to be decondensed to a nucleosome structural level in order to transcribe genes [13, 14]. The length of the decondensed chromatin loop can sometimes exceed the area occupied by the chromosome territory, to which the loop belongs, allowing it to intermingle into the neighbouring chromosome territory [15].
Chromosome and chromosome territories[Edit]
While metaphase chromosomes can be depicted as distinct bodies with well-defined shapes and sizes, interphase chromosomes are less uniform and, by filling the nuclear space, difficult to distinguish. Despite this, recent research has revealed how the nuclear architecture dictates interphase chromosome arrangement and territorial organization in differentiated cells.
 Figure 3. From DNA to metaphase chromosome: Folding of DNA into nucleosomes achieves initial 6-fold compaction level. Histone variants present in the nucleosome core, posttranslational modifications and linker histone H1 position can all control DNA accessibility for transcription at this compaction level. Further chromatin condensation into 30 nm fibers (i.e., zigzag or solenoid) is suggested by in vitro data and is yet to be proved or discredited to exist in vivo. During the interphase, chromatin is folded into 300-700 nm domains, which together comprise a chromosome territory. The structure and organization of chromatin loops inside a chromosome territory remains the matter of debates and was proposed to exist in the form of solenoid, or zigzag, or nucleosomes, or a hybrid of those.During interphase, each chromosome occupies a spatially limited, roughly elliptical domain which is known as a chromosome territory (CT) [16, 17]. Each chromosome territory is comprised of higher order chromatin units of ~1 Mb each. These units are likely built up from smaller loop domains. On the other hand, 1Mb domains can themselves serve as smaller units in higher-order chromatin structures [16].
Figure 3. From DNA to metaphase chromosome: Folding of DNA into nucleosomes achieves initial 6-fold compaction level. Histone variants present in the nucleosome core, posttranslational modifications and linker histone H1 position can all control DNA accessibility for transcription at this compaction level. Further chromatin condensation into 30 nm fibers (i.e., zigzag or solenoid) is suggested by in vitro data and is yet to be proved or discredited to exist in vivo. During the interphase, chromatin is folded into 300-700 nm domains, which together comprise a chromosome territory. The structure and organization of chromatin loops inside a chromosome territory remains the matter of debates and was proposed to exist in the form of solenoid, or zigzag, or nucleosomes, or a hybrid of those.During interphase, each chromosome occupies a spatially limited, roughly elliptical domain which is known as a chromosome territory (CT) [16, 17]. Each chromosome territory is comprised of higher order chromatin units of ~1 Mb each. These units are likely built up from smaller loop domains. On the other hand, 1Mb domains can themselves serve as smaller units in higher-order chromatin structures [16].
Chromosome territories are known to be arranged radially around the nucleus. This arrangement is both cell and tissue-type specific and is also evolutionary conserved [18].
 Figure 4. Possible arrangements of chromosome territories (CTs) and interchromatin compartment (IC).A. Chromosome territory – interchromatin compartment model (CT-IC). B. Interchromatin network model (ICN).: On the CT-IC model, the space between discrete CTs can be visualized in light and electron microscope and is called interchromatin compartment (IC). Transcription factories (TF, green color) are localized predominantly in perichromatin region. In the ICN model, interchromatin compartment is not apparent. Instead, the space between CTs is occupied by intermingling decondensed chromatin loops, which often share the same transcription factories.
Within a single chromosome territory, the interphase chromosome is divided into defined regions based on the level of chromosome condensation. Here, the inner part of the interphase chromosome is comprised of more condensed chromatin domains or higher-order chromatin fibers, while a thin (<200 nm) layer of more decondensed chromatin, known as the perichromatin region, can be found around the chromosomal periphery [30]. Functionally, the perichromatin region represents the major transcriptional compartment, and is also the region where most co-transcriptional RNA splicing takes place [31, 32, 33]. DNA replication [34] and DNA repair [35] is also predominately carried out within the perichromatin region. Finally, nascent RNA transcripts, referred to as perichromatin fibrils, are also generated in the perichromatin region. Perichromatin fibrils are then subjected to the splicing events by the factors, provided from the interchromatin compartment.
Figure 4. Possible arrangements of chromosome territories (CTs) and interchromatin compartment (IC).A. Chromosome territory – interchromatin compartment model (CT-IC). B. Interchromatin network model (ICN).: On the CT-IC model, the space between discrete CTs can be visualized in light and electron microscope and is called interchromatin compartment (IC). Transcription factories (TF, green color) are localized predominantly in perichromatin region. In the ICN model, interchromatin compartment is not apparent. Instead, the space between CTs is occupied by intermingling decondensed chromatin loops, which often share the same transcription factories.
Within a single chromosome territory, the interphase chromosome is divided into defined regions based on the level of chromosome condensation. Here, the inner part of the interphase chromosome is comprised of more condensed chromatin domains or higher-order chromatin fibers, while a thin (<200 nm) layer of more decondensed chromatin, known as the perichromatin region, can be found around the chromosomal periphery [30]. Functionally, the perichromatin region represents the major transcriptional compartment, and is also the region where most co-transcriptional RNA splicing takes place [31, 32, 33]. DNA replication [34] and DNA repair [35] is also predominately carried out within the perichromatin region. Finally, nascent RNA transcripts, referred to as perichromatin fibrils, are also generated in the perichromatin region. Perichromatin fibrils are then subjected to the splicing events by the factors, provided from the interchromatin compartment.
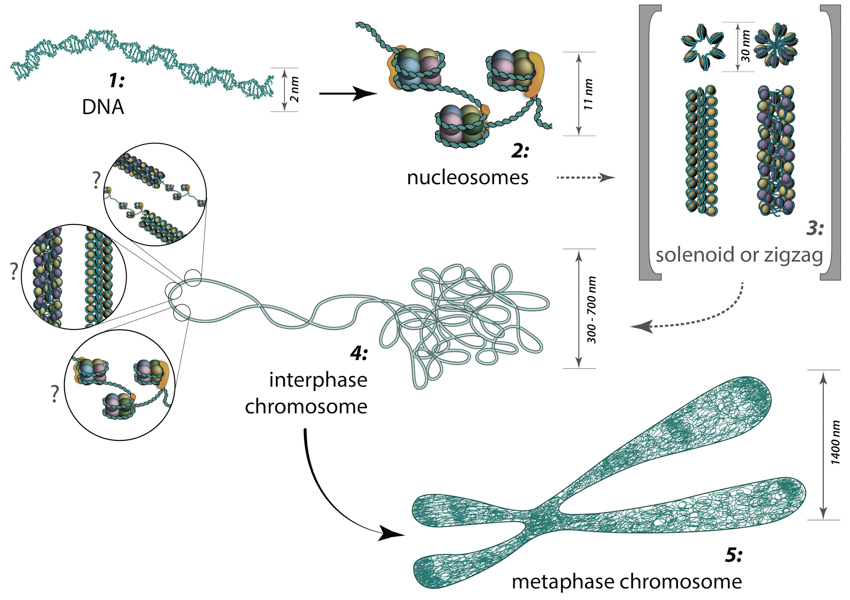 Figure 3. From DNA to metaphase chromosome: Folding of DNA into nucleosomes achieves initial 6-fold compaction level. Histone variants present in the nucleosome core, posttranslational modifications and linker histone H1 position can all control DNA accessibility for transcription at this compaction level. Further chromatin condensation into 30 nm fibers (i.e., zigzag or solenoid) is suggested by in vitro data and is yet to be proved or discredited to exist in vivo. During the interphase, chromatin is folded into 300-700 nm domains, which together comprise a chromosome territory. The structure and organization of chromatin loops inside a chromosome territory remains the matter of debates and was proposed to exist in the form of solenoid, or zigzag, or nucleosomes, or a hybrid of those.
Figure 3. From DNA to metaphase chromosome: Folding of DNA into nucleosomes achieves initial 6-fold compaction level. Histone variants present in the nucleosome core, posttranslational modifications and linker histone H1 position can all control DNA accessibility for transcription at this compaction level. Further chromatin condensation into 30 nm fibers (i.e., zigzag or solenoid) is suggested by in vitro data and is yet to be proved or discredited to exist in vivo. During the interphase, chromatin is folded into 300-700 nm domains, which together comprise a chromosome territory. The structure and organization of chromatin loops inside a chromosome territory remains the matter of debates and was proposed to exist in the form of solenoid, or zigzag, or nucleosomes, or a hybrid of those.Chromosome territories are known to be arranged radially around the nucleus. This arrangement is both cell and tissue-type specific and is also evolutionary conserved [18].
The radial organization of chromosome territories was shown to correlate with their gene density and size. In this case, the gene-rich chromosomes occupy interior positions, whereas larger, gene-poor chromosomes, tend to be located around the periphery [19, 20, 21]. Chromosome territories are also dynamic structures, with genes able to relocate from the periphery towards the interior once they have been “switched on”[22]. In other cases, genes may move in the opposite direction, or simply maintain their position [23,24]. The eviction of genes from their chromosome territories into the interchromatin compartment or a neighbouring chromosome territory is often accompanied by the formation of large decondensed chromatin loops[18].
With the development of high-throughput biochemical techniques, such as 3C (“chromosome conformation capture”)[25] and 4C (“chromosome conformation capture-on-chip” [26] and “circular chromosome conformation capture” [27]), numerous spatial interactions between neighbouring chromatin territories have been described. These descriptions have been supplemented with the construction of spatial proximity maps for the entire genome (e.g., for a human lymphoblastoid cell line [12]). Together, these observations and physical simulations have led to the proposal of various models that aim to define the structural organization of chromosome territories [16]:
1. The chromosome territory-interchromatin compartment (CT-IC) model describes two principal compartments: chromosome territories (CTs) and an interchromatin compartment (IC). In this model, chromosome territories build up an interconnected chromatin network [28] that is associated with an adjacent 3D space called the interchromatin compartment. The latter can be observed using both light and electron microscopy [29]
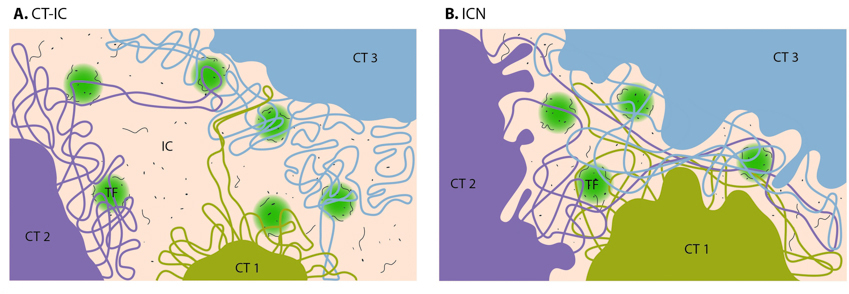 Figure 4. Possible arrangements of chromosome territories (CTs) and interchromatin compartment (IC).A. Chromosome territory – interchromatin compartment model (CT-IC). B. Interchromatin network model (ICN).: On the CT-IC model, the space between discrete CTs can be visualized in light and electron microscope and is called interchromatin compartment (IC). Transcription factories (TF, green color) are localized predominantly in perichromatin region. In the ICN model, interchromatin compartment is not apparent. Instead, the space between CTs is occupied by intermingling decondensed chromatin loops, which often share the same transcription factories.
Figure 4. Possible arrangements of chromosome territories (CTs) and interchromatin compartment (IC).A. Chromosome territory – interchromatin compartment model (CT-IC). B. Interchromatin network model (ICN).: On the CT-IC model, the space between discrete CTs can be visualized in light and electron microscope and is called interchromatin compartment (IC). Transcription factories (TF, green color) are localized predominantly in perichromatin region. In the ICN model, interchromatin compartment is not apparent. Instead, the space between CTs is occupied by intermingling decondensed chromatin loops, which often share the same transcription factories.The lattice model, proposed by Dehgani et al. [36] is based on reports that transcription also occurs within the inner, more condensed chromosome territories and not only at the interface between the interchromatin compartment and the perichromatin region [37, 38, 39, 40]. Using ESI (electron spectroscopic imaging), Dehgani et al. showed that chromatin was organized as an array of deoxy-ribonucleoprotein fibers of 10–30 nm in diameter. In this study, the interchromatin compartments, which are described in the CT-IC model as large channels between chromosome territories, were not apparent. Instead, chromatin fibers created a loose meshwork of chromatin throughout the nucleus that intermingled at the periphery of chromosome territories. Thus, inter- and intra-chromosomal spaces within this meshwork are essentially contiguous and together form the intra-nuclear space [36].
2. The interchromatin network (ICN) model [41] predicts that intermingling chromatin fibers/loops can make both cis- (within the same chromosome) and trans- (between different chromosomes) contacts. This intermingling is uniform and makes distinction between the chromosome territory and interchromatin compartment functionally meaningless [16]. The advantage of the ICN model is that it permits high chromatin dynamics and diffusion-like movements. The authors propose that ongoing transcription influences the degree of intermingling between specific chromosomes by stabilizing associations between particular loci. Such interactions are likely to depend on the transcriptional activity of the loci, and are therefore cell-type specific.
3. The Fraser and Bickmore model [42] emphasizes the functional importance of giant chromatin loops, which originate from chromosome territories and expand across the nuclear space in order to share transcription factories. In this case, both cis- and trans- loops of decondensed chromatin can be co-expressed and co-regulated by the same transcription factory.
4. The Chromatin polymer models assume a broad range of chromatin loop sizes [43] and predict the observed distances between genomic loci and chromosome territories, as well as the probabilities of contacts being formed between given loci [44]. These models apply physics-based approaches that highlight the importance of entropy for understanding nuclear organization. By proposing the existence of conformational chromatin ensembles with structures based on three possible homopolymer states, these models also provide alternative structures to the traditional 30 nm chromatin fiber, which has been brought into question following recent studies [45, 46, 47].
With a lack of experimental evidence to support these described models, it must be remembered that they serve only to hypothesize the structural and chemical properties of intermediate chromatin structures, and to highlight unanswered questions [16]. For example, the mechanisms that exist to control the rate and the extent of chromatin movement remain to be defined