Actin Filament
Content
- What are actin filaments
- Actin Filament Polarity
- Distribution of actin filaments in cells and tissues
- Actin Filament Function
- Actin filament polymerization generates protrusive force
- Actin filaments are a force-sensing conduit for both internal and external forces
- Actin filaments form higher-order assemblies that produce and respond to force
What are actin filaments[Edit]
Actin filaments (F-actin) are linear polymers of globular actin (G-actin) subunits and occur as microfilaments in the cytoskeleton and as thin filaments, which are part of the contractile apparatus, in muscle and nonmuscle cells (see contractile bundles). They commonly underlie the plasma membrane and are typically assembled at the cell periphery from adhesion sites or sites of membrane extension. Actin filaments can create a number of linear bundles, two-dimensional networks, and three-dimensional gels, and actin binding proteins can influence the specific structure the filaments will form.
Although a large body of work suggests that F-actin can exist in multiple states, in general, an actin filament has a total rise of 27.3 Å between subunits on adjacent strands and a rotation of 166.15° around the axis [1]. An actin filament is flexible, has a helical repeat every 37 nm, ranges from 5-9 nm in diameter, and has 13 actin subunits between each cross-over point (produced by the ‘crossing over’ of the two long-pitch actin helices) (reviewed in [2, 3]). An exception is the case of sperm, where a crosslinker protein, scruin, maintains the actin in a coiled state and extends upon activation. In the acrosomal actin rotation angle of one of the strands is 167.15° while the other strand is tilted more than usual to fit into an asymmetric helix [4].
Actin Filament Polarity[Edit]
The polarity of an actin filament is visualized by the binding of the myosin subfragment (S1) to the filament, which creates barbed (+) and pointed (-) ends on the filament [5].  Figure 1. Structure of an actin filament showing the barbed (or plus) and pointed (or minus) ends: This diagram illustrates the molecular organization of actin and provides examples for how an actin filament is represented in figures throughout this resource. Early models for actin filaments were constructed by fitting the filament x-ray crystal structure to the atomic structure of actin monomers [6](reviewed in [2]) while more recent models use a number of different approaches [7].When all actin subunits are bound by myosin S1, the filament appears coated with arrowheads that all point towards one end of the filament [8, 9]. The barbed end structure with bound capping protein has been determined [10] and polarity ascertained by novel single-particle image analysis methods applied to electromicrographs of in vitro protein preparations [11]. Polarity has since also been determined from cryo-electron tomograms of vitreously frozen cells [12]. The (+) end of filaments generally have a high concentration of F-actin-ATP and denotes the growing end of an actin filament.
Figure 1. Structure of an actin filament showing the barbed (or plus) and pointed (or minus) ends: This diagram illustrates the molecular organization of actin and provides examples for how an actin filament is represented in figures throughout this resource. Early models for actin filaments were constructed by fitting the filament x-ray crystal structure to the atomic structure of actin monomers [6](reviewed in [2]) while more recent models use a number of different approaches [7].When all actin subunits are bound by myosin S1, the filament appears coated with arrowheads that all point towards one end of the filament [8, 9]. The barbed end structure with bound capping protein has been determined [10] and polarity ascertained by novel single-particle image analysis methods applied to electromicrographs of in vitro protein preparations [11]. Polarity has since also been determined from cryo-electron tomograms of vitreously frozen cells [12]. The (+) end of filaments generally have a high concentration of F-actin-ATP and denotes the growing end of an actin filament.
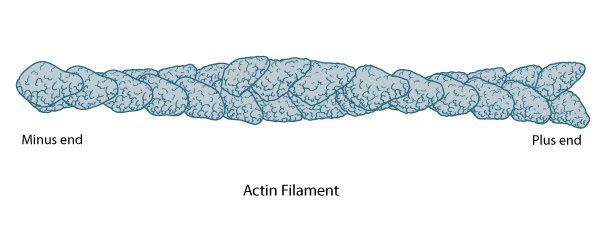 Figure 1. Structure of an actin filament showing the barbed (or plus) and pointed (or minus) ends: This diagram illustrates the molecular organization of actin and provides examples for how an actin filament is represented in figures throughout this resource. Early models for actin filaments were constructed by fitting the filament x-ray crystal structure to the atomic structure of actin monomers [6](reviewed in [2]) while more recent models use a number of different approaches [7].
Figure 1. Structure of an actin filament showing the barbed (or plus) and pointed (or minus) ends: This diagram illustrates the molecular organization of actin and provides examples for how an actin filament is represented in figures throughout this resource. Early models for actin filaments were constructed by fitting the filament x-ray crystal structure to the atomic structure of actin monomers [6](reviewed in [2]) while more recent models use a number of different approaches [7].The (-) end generally has a high concentration of F-actin-ADP and denotes the shrinking end of an actin filament. Most actin filaments are arranged with the barbed end toward the cellular membrane and the pointed end toward the cellular interior. The length of actin filaments can be adjusted by the activity of actin binding proteins and by altering the rate of actin treadmilling.
Distribution of actin filaments in cells and tissues[Edit]
Actin filaments are widely distributed throughout cells, forming a range of cytoskeletal structures and contributing to an even broader range of processes. Some of the functions are widely observed across many cell types while in other cases actin filaments may contribute to processes that are cell type specific.
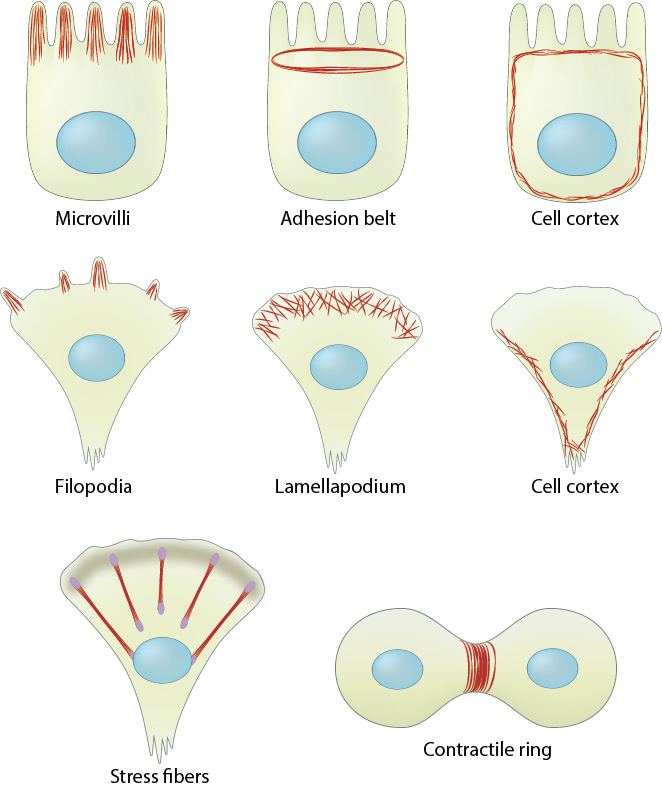
Actin filaments are crucial for tissue organization and for establishing cell polarity and cohesion among epithelial cells. For example, a core of actin filaments provides microvilli structural support and enables them to increase their surface area and nutrient-absorbing capacity. These structures are found on the apical surface of epithlial cells lining the small intestine. In another example, the integrity of epithelial cell layers or sheets is maintained by a belt of actin filaments (i.e. adhesion belt). This belt links the cytoskeleton of adjacent cells. Also, certain cells use actin filament rigidity to sense vibrations, such as those found bundled on the surface of hair cells in the inner ear (called stereocilia, not shown), which tilt in response to sound. Although the actin bundles in stereocilia are stable for the lifetime of a cell (which can be decades), the individual actin filaments are continuously remodeled and replaced once every 48 hours (on average).
 Figure 3. Cell cortex (aka cortical actin, actin cortex): (A) Cortical actin filaments (shown in red) are concentrated just beneath the plasma membrane in most cell types. (B) Migrating fibroblasts grown in 2D tissue culture have more cortical filaments on the dorsal (upper) surface than the ventral (lower) surface and they are concentrated towards the trailing edges [13]
Actin filaments are the primary cytoskeletal component to drive cell motility. Here, actin filaments found in membrane protrusions such as filopodia and lamellipodia rapidly assemble and disassemble. These cellular structures are essential in cell migration and are predominately found at the leading edge of a moving cell. They also allow the cell to probe or sense its microenvironment. Actin filaments are also found at the trailing edge in the form of the cell cortex, which lies adjacent to the plasma membrane. More stable arrays of actin filaments, such as those found in stress fibers, allow a cell to brace against the underlying surface. Actin-associated myosin motor proteins use ATP hydrolysis to exert forces against the stress fibers during muscle contraction; the energy of hydrolysis can also be converted to tensile forces at the trailing cell edge to promote edge retraction in motile cells.
Figure 3. Cell cortex (aka cortical actin, actin cortex): (A) Cortical actin filaments (shown in red) are concentrated just beneath the plasma membrane in most cell types. (B) Migrating fibroblasts grown in 2D tissue culture have more cortical filaments on the dorsal (upper) surface than the ventral (lower) surface and they are concentrated towards the trailing edges [13]
Actin filaments are the primary cytoskeletal component to drive cell motility. Here, actin filaments found in membrane protrusions such as filopodia and lamellipodia rapidly assemble and disassemble. These cellular structures are essential in cell migration and are predominately found at the leading edge of a moving cell. They also allow the cell to probe or sense its microenvironment. Actin filaments are also found at the trailing edge in the form of the cell cortex, which lies adjacent to the plasma membrane. More stable arrays of actin filaments, such as those found in stress fibers, allow a cell to brace against the underlying surface. Actin-associated myosin motor proteins use ATP hydrolysis to exert forces against the stress fibers during muscle contraction; the energy of hydrolysis can also be converted to tensile forces at the trailing cell edge to promote edge retraction in motile cells.
2. In motility-related structures:
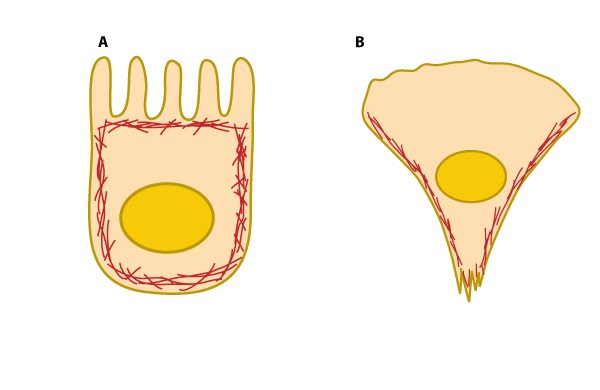 Figure 3. Cell cortex (aka cortical actin, actin cortex): (A) Cortical actin filaments (shown in red) are concentrated just beneath the plasma membrane in most cell types. (B) Migrating fibroblasts grown in 2D tissue culture have more cortical filaments on the dorsal (upper) surface than the ventral (lower) surface and they are concentrated towards the trailing edges [13]
Figure 3. Cell cortex (aka cortical actin, actin cortex): (A) Cortical actin filaments (shown in red) are concentrated just beneath the plasma membrane in most cell types. (B) Migrating fibroblasts grown in 2D tissue culture have more cortical filaments on the dorsal (upper) surface than the ventral (lower) surface and they are concentrated towards the trailing edges [13]Actin-based motile structures are disassembled before cell division, which causes the cell to stop moving and become more rounded. More stable actin bundles remain polarized and contribute to the orientation of the microtubule network that serves as the mitotic spindle. The proper assembly, orientation, and contraction of an actin filament ring (i.e. contractile ring) serves to pinch and separate the daughter cells during cell division.
4. During reproduction:
Actin filaments are an important structural feature in the head of sperm. The rapid relaxing of coiled actin filaments during the acrosome reaction [4] allows the sperm head to penetrate further into the egg. Microtubules influence actin assembly to help organize the polarized actin network.Actin Filament Function[Edit]
Several biological processes related to cell shape and movement depend on actin filaments (reviewed in [14]). Some keys functions are:
* To form the dynamic cytoskeleton, which gives structural support to cells and links the interior of the cell with its surroundings. Forces acting on the actin cytoskeleton are translated and transmitted by signaling pathways to convey information about the external environment.
* To form the dynamic cytoskeleton, which gives structural support to cells and links the interior of the cell with its surroundings. Forces acting on the actin cytoskeleton are translated and transmitted by signaling pathways to convey information about the external environment.
* To allow cell motility. For example, through the formation and function of Filopodia or Lamellipodia.
* During mitosis, intracellular organelles are transported by motor proteins to the daughter cells along actin cables
* In muscle cells, actin filaments are aligned and myosin proteins generate forces on the filaments to support muscle contraction. These complexes are known as ‘thin filaments’.
* In non-muscle cells, actin filaments form a track system for cargo transport that is powered by non-conventional myosins such as myosin V and VI. Non-conventional myosins use the energy from ATP hydrolysis to transport cargo (such as vesicles and organelles) at rates much faster than diffusion.
* In muscle cells, actin filaments are aligned and myosin proteins generate forces on the filaments to support muscle contraction. These complexes are known as ‘thin filaments’.
* In non-muscle cells, actin filaments form a track system for cargo transport that is powered by non-conventional myosins such as myosin V and VI. Non-conventional myosins use the energy from ATP hydrolysis to transport cargo (such as vesicles and organelles) at rates much faster than diffusion.
Thin filament
In certain cases, actin filaments are assembled with, and stabilized by, accessory proteins into higher order contractile structures such as stress fibers (nonmuscle cells) or contractile bundles (muscle cells). The dynamic association of tropomyosin and troponin with actin filaments stabilizes the actin filament (collectively termed “thin filament”) to be functional in various contexts.
 Figure 4. Tropomyosin stabilizes thin filaments: TM binds to the side of adjacent actin subunits along the groove of the helix to stabilize and stiffen the actin filament [15]. TM also prevents other proteins from accessing the filament; this inhibition is essential for regulating muscle contraction [16]. TN controls the positioning of TM along the groove of the actin filament.A single tropomyosin binds to the side of adjacent actin subunits and extends over approximately seven actin monomers [15]. End-to-end binding of tropomyosins produces a continuous strand of tropomyosin polymers along the groove of the actin helix which allows their cooperative movement [17]. Tropomyosin isoforms stabilize actin filaments [18] and occupy the same binding sites on actin that are used by known regulators of actin filaments (e.g. ADF/cofilin [19]) (reviewed in [20, 21]).
Figure 4. Tropomyosin stabilizes thin filaments: TM binds to the side of adjacent actin subunits along the groove of the helix to stabilize and stiffen the actin filament [15]. TM also prevents other proteins from accessing the filament; this inhibition is essential for regulating muscle contraction [16]. TN controls the positioning of TM along the groove of the actin filament.A single tropomyosin binds to the side of adjacent actin subunits and extends over approximately seven actin monomers [15]. End-to-end binding of tropomyosins produces a continuous strand of tropomyosin polymers along the groove of the actin helix which allows their cooperative movement [17]. Tropomyosin isoforms stabilize actin filaments [18] and occupy the same binding sites on actin that are used by known regulators of actin filaments (e.g. ADF/cofilin [19]) (reviewed in [20, 21]).
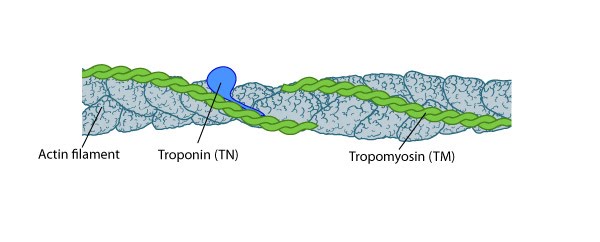 Figure 4. Tropomyosin stabilizes thin filaments: TM binds to the side of adjacent actin subunits along the groove of the helix to stabilize and stiffen the actin filament [15]. TM also prevents other proteins from accessing the filament; this inhibition is essential for regulating muscle contraction [16]. TN controls the positioning of TM along the groove of the actin filament.
Figure 4. Tropomyosin stabilizes thin filaments: TM binds to the side of adjacent actin subunits along the groove of the helix to stabilize and stiffen the actin filament [15]. TM also prevents other proteins from accessing the filament; this inhibition is essential for regulating muscle contraction [16]. TN controls the positioning of TM along the groove of the actin filament.Focus on contractile machinery
Troponin, a three-peptide complex, is thought to trap tropomyosin in a calcium-dependent fashion at a position that inhibits myosin bundles from accessing the actin filaments; calcium binding to troponin allows a conformational restructuring of tropomyosin that leaves the myosin-binding sites on the thin filaments exposed [22, 23, 24, 25]. Subsequent binding of the myosin thick filaments augments movement of tropomyosin away from the actin filament and full exposure of the myosin binding sites [24]. However, control of tropomyosin-binding to myosin thick filaments may be independent of troponin presence [18]; smooth muscle cells and many non-muscle cells lack troponin.Actin filament polymerization generates protrusive force[Edit]
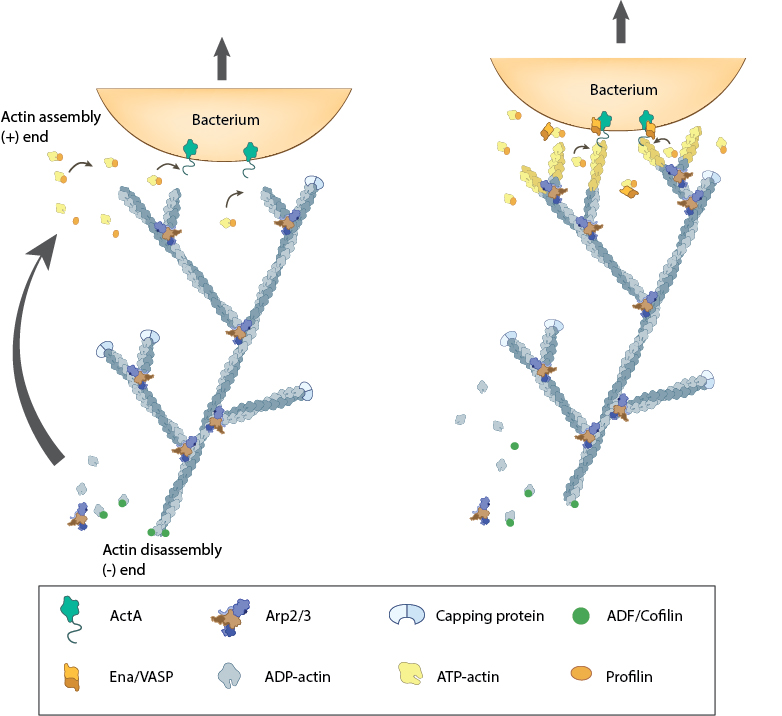 Figure 5. Actin polymerization produces force for movement: Pathogenic intracellular bacteria use mainly host-derived components to facilitate movement through (and between) cells. The bacterial ActA protein on the surface of the bacterium (e.g. Listeria monocytogenes) recruits and activates the host components needed for actin polymerization (e.g. Ena/VASP, ATP-actin). The actin filaments are assembled at the plus end nearest the bacteria membrane and the forces generated by filament assembly are translated into bacterium movement. The resulting actin comet tail is likely comprised of branched arrays of actin filaments as seen from simulation of experimental results [28, 29, 30]. The dynamics of filament assembly and turnover (and therefore bacterium motility) are controlled by capping protein, profilin, ADF/Cofilin and Arp2/3 complex.
Figure 5. Actin polymerization produces force for movement: Pathogenic intracellular bacteria use mainly host-derived components to facilitate movement through (and between) cells. The bacterial ActA protein on the surface of the bacterium (e.g. Listeria monocytogenes) recruits and activates the host components needed for actin polymerization (e.g. Ena/VASP, ATP-actin). The actin filaments are assembled at the plus end nearest the bacteria membrane and the forces generated by filament assembly are translated into bacterium movement. The resulting actin comet tail is likely comprised of branched arrays of actin filaments as seen from simulation of experimental results [28, 29, 30]. The dynamics of filament assembly and turnover (and therefore bacterium motility) are controlled by capping protein, profilin, ADF/Cofilin and Arp2/3 complex.In cells, actin filaments are initiated with their barbed ends oriented towards the plasma membrane, with ATP hydrolysis facilitating filament growth. Polymerization is favored towards the cell front and disassembly occurs more frequently at the rear (reviewed in [39]). However, only a small fraction of the overall free energy of nucleotide hydrolysis is needed to modulate G-actin monomer binding. The remaining energy is translated into a protrusive force that deforms the plasma membrane in a particular direction [40, 41, 42, 43] (reviewed in [35]).
The propulsive network is self-organizing and filaments with a particular orientation,with respect to the membrane, will assemble at the maximal velocity and be preferentially chosen for elongation [44]. Similarly, cell shape and migration speed is determined by a dynamic steady state that is self-organizing [45]. As actin filaments grow, they remain fixed within the cytoskeletal network (reviewed in [46]).
Factors that regulate the protrusive force of actin filaments:
Factors that influence the concentration of free G-actin (e.g. profilin [47]) or ATP-binding and hydrolysis on actin (reviewed in [38]) will promote filament assembly and membrane protrusion.The microtubule and intermediate filament networks play a key role in regulating the global deposition pattern of the actin filaments, therefore, they will also influence membrane protrusion dynamics [48] (reviewed in [44, 45, 49, 50, 51]).
Membrane tension- In order for a cell to extend its leading edge forward, the cell must overcome resisting forces. Motile cells in living systems experience external forces from the surrounding material (usually ECM), while the major force resisting extension for cells in tissue culture are tensile forces within the plasma membrane. Biophysical models [44, 52, 53] (reviewed in [54, 55]) and experiments with live cells have shown that the membrane extension rate is directly dependent on the membrane tension: elevated tension lowers cell membrane extension and motility, regardless of whether the tension is applied externally (e.g. stretching) or internally (e.g. contraction of stress fibers) [43, 56, 57].
Actin filaments are a force-sensing conduit for both internal and external forces[Edit]
Internal forces
The orientation of individual actin filaments in the cytoskeleton is a force-driven evolutionary process [58] that contributes to the elastic behavior of the network and influences whether a filament will deform by compression, bending or extension [33, 59].Cross-linked actin networks initially become more elastic under low force as a result of filament resistance to the direction of compression [60]. As the force increases, individual filaments inherently resist being compressed and/or cross-linking proteins become more extended, which causes the cytoskeleton network to become more rigid [61, 62]; cell stiffening has also been correlated with actin recruitment [63]. Each of these processes in turn, can influence mechanosensors that are linked to the actin network to induce subsequent mechanotransduction events. Although forces ~ 400 pN along the filament axis will break actin filaments, much higher forces are generated in migrating cells (reviewed in[64]); this raises the question “how does a cell alleviate the forces on the internal actin network?” The answer lies in the proteins that are linked to the actin: extreme stress on the filaments causes buckling and reduced elasticity of the network in a process known as ‘stress softening’ [65]. Softening is attributed to either filament fracturing, which leads to new uncapped ends for filament assembly/disassembly, or to softening, which may be due to unbinding of flexible protein crosslinkers [60, 66]. In addition, because the filaments are interconnected to the rest of the cytoskeleton, buckled filaments don’t collapse and the process can be reversed when the stress is removed [67] (reviewed in [68, 69]).
External or applied forces
The actin cytoskeleton is physically connected with the cell exterior, e.g., ECM or other cells, through a multiprotein complex known as the focal adhesion complex. Interactions between cell surface molecules, e.g., integrins, and the actin cytoskeleton are bidirectional, with the focal adhesion complex forming the link between them [70]. Actin filaments and their associated focal adhesion complexes act as information handling machines or mechanosensors: they convert both the strength of the adhesion and the tensile forces along the linked network of actin filaments (and associated proteins) into biochemical signals that control actin extension and cell migration (reviewed in [69, 71, 72,73, 74, 75]). Focal adhesions are subject to continuous pulling forces [76, 77] and the force differences due to external stress [78] or the chemical nature (reviewed in [79]), rigidity [80,81], and topography of the exterior components [82, 83, 84] (reviewed in [46, 85, 86]) will influence the assembly and organization of the actin cytoskeleton [84, 45, 87, 88, 89].
Actin filaments form higher-order assemblies that produce and respond to force[Edit]
Many different types of cells use actin filaments to generate tensional forces and to exert traction forces on their adhesions and linked ECM (or other cells); these processes cause resting tensile force (reviewed in [68, 90]). Local stress or force differences that occur both internally and externally are transduced through the actin filaments and focused onto mechanosensors throughout the interlinked system rather than being limited to local deformation (reviewed in [91]); this process leads to mechanotransduction events that influence the cell shape and/or motility (reviewed in [92]).
Cells exert traction forces on the ECM and generate tension at focal adhesions through actin stress fibers, which are higher-order structures in the cytoplasm that consist of parallel contractile bundles of actin and myosin filaments. Stress fibers are linked at their ends to the ECM through focal adhesion complexes. Cell tension is generated along the actin filaments by the movement of myosin II motor proteins along the filaments (see contractile bundles). The elasticity of the actin network appears to be inherent to actin filaments and is independent of myosin II motor activity [65]. Forces produced by the contraction of stress fibers not only helps the cell body to translocate during migration [84, 93], but they also serve as a vital “inside-out” feedback system to regulate actin filament initiation [94], cell growth and motility [95, 96], and formation/maturation of focal adhesion complexes [97, 98]. Forces produced by stress fibers also stabilize the cell structure and contribute to establishing the cell polarity [93] and they help determine the cell fate [76, 99]. There is no evidence that forces influence stress fiber contractile activity by increasing the exchange of ADP for ATP on myosin, however, force can weakly increase the release of the myosin heads from the actin filaments (reviewed in [64]).
Cells exert traction forces on the ECM and generate tension at focal adhesions through actin stress fibers, which are higher-order structures in the cytoplasm that consist of parallel contractile bundles of actin and myosin filaments. Stress fibers are linked at their ends to the ECM through focal adhesion complexes. Cell tension is generated along the actin filaments by the movement of myosin II motor proteins along the filaments (see contractile bundles). The elasticity of the actin network appears to be inherent to actin filaments and is independent of myosin II motor activity [65]. Forces produced by the contraction of stress fibers not only helps the cell body to translocate during migration [84, 93], but they also serve as a vital “inside-out” feedback system to regulate actin filament initiation [94], cell growth and motility [95, 96], and formation/maturation of focal adhesion complexes [97, 98]. Forces produced by stress fibers also stabilize the cell structure and contribute to establishing the cell polarity [93] and they help determine the cell fate [76, 99]. There is no evidence that forces influence stress fiber contractile activity by increasing the exchange of ADP for ATP on myosin, however, force can weakly increase the release of the myosin heads from the actin filaments (reviewed in [64]).