Contents Quiz Test Your Knowledge! Click Here Contribute | Essential Info: Actin Filaments and Mechanotransduction4.3 Dynamics of actin filament assembly and treadmillingActin filaments are dynamic structures whose growth and shrinkage are tightly controlled. Their nucleation occurs primarily at or near the plasma membrane. Consequently a region of high actin filament density is commonly found at the cell periphery – this is called the cell cortex. Actin filaments in the cell cortex determine the shape, stiffness and movement of the cell surface. Filament assembly requires a set of minimal components (as described below), which promote actin filament nucleation and can lead to a steady state process termed actin treadmilling.Minimal Components for Actin Filament AssemblyActin monomers are globular proteins that bind to ATP or ADP. ATP hydrolysis on these monomers alters their conformation and so influences actin filament dynamics. During the initial phase of actin filament assembly, actin monomers will spontaneously oligomerize in solution when present at a concentration above the critical concentration (Cc) (see Table 1 and Figure below), but these complexes are highly unstable.Efficient nucleation and assembly of actin filament networks that drive the motility of pathogenic, intracellular bacteria, such as Listeria, requires the minimum components of actin, Arp2/3 complex, profilin, ADF/cofilin and capping protein [1]. This is true for most other eukaryotic cells, though quantification has only been carried out in a limited number of organisms (see Table 2) [2]. The cooperation between each component is extensive and each element has an optimal concentration. If the pool of actin monomers is large, actin filament assembly eventually reaches a steady state and the actin filaments appear to ‘treadmill’ [3].
Table 1: Actin association and dissociation rates on actin filaments (reviewed in [4]).
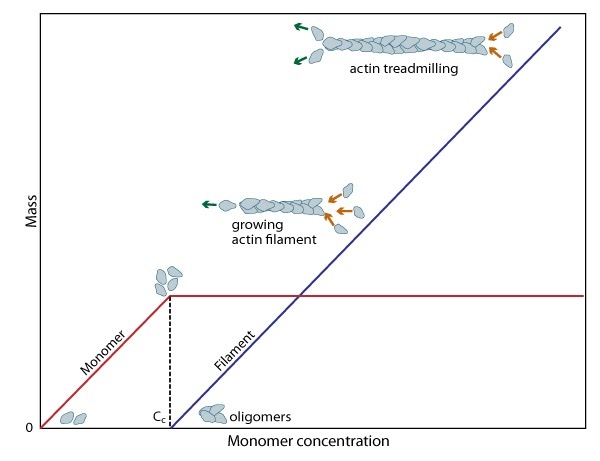
Figure: Cc and actin filament assembly. The critical concentration (Cc)
marks the level at which G-actin monomers are in equilibrium with the
actin filaments. Actin filaments are only formed at monomer
concentrations above the Cc.
Table 2: Cellular concentrations (in µM) of key proteins in the actin system of diverse cells (if known, modified from [2] and [5]). * These values are representative of biological concentrations similar to those used for in vitro reconstitution experiments of bacterial motility and so are not cell type specific [5].
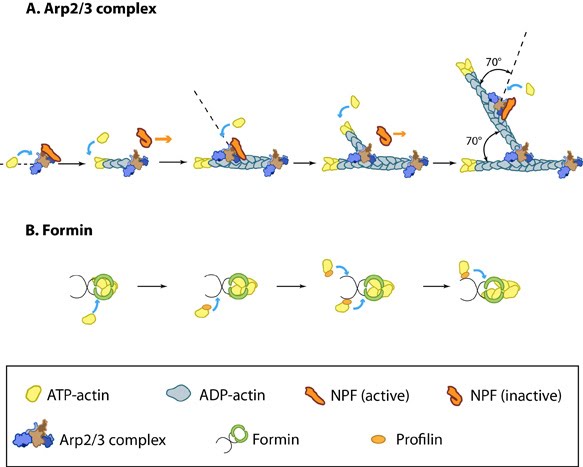
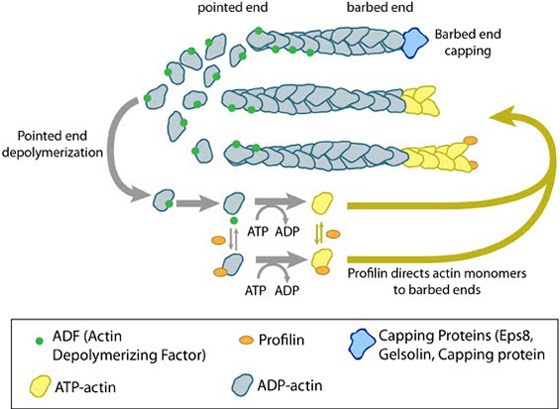
Actin Filament NucleationThe nucleation and/or branching of new actin filaments and their assembly are critical for cellular function, in order to facilitate changes in cell motility, structure and stiffness. Most ATP-actin polymerization occurs at the barbed end of an actin filament. The barbed ends are created by uncapping or severing existing filaments or by de novo nucleation.Filament nucleation is controlled by both the accessibility of barbed ends and by factors that bind monomeric actin and modulate its polymerization properties. A steady pool of actin monomers is maintained by profilin, which promotes ADP to ATP nucleotide exchange on G-actin. Although nonmuscle cells have a high concentration of G-actin-ATP (~100 μM) [6], pure G-actin monomers fail to nucleate new actin filaments efficiently due to the instability of actin oligomers. Additional factors are therefore necessary for the production of actin filaments. The Arp2/3 complex and formins are among the most widely studied proteins which regulate actin nucleation and assembly (For more information, see the functional modules Arp2/3 complex and Formin and Profilin in Actin Nucleation). Figure: Accessory proteins nucleate actin filaments. A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament. Actin TreadmillingFollowing actin nucleation, filaments are able to assemble. When the association rate of free ATP-G-actin to the ends of actin filaments is greater than the rate of subunit loss, the filament is seen to grow. Conversely, when the association rate of free ATP-actin to the ends of actin filaments is lower than the rate of subunit loss, the filament is seen to shrink. Actin treadmilling occurs when the association rate of free ATP-actin to the ends of actin filaments is balanced by the rate of subunit loss and no net growth occurs. Actin treadmilling is powered by ATP hydroylsis (as reviewed in [7]) and this energy can be used to perform work. For more information on the regulation of this process, see Factors influencing actin filament length and treadmilling.Figure: Regulation of actin treadmilling. The length of actin filaments is controlled by actin binding proteins. Capping proteins prevent assembly at the barbed end while ADF binds to the side of ADP-actin filaments to cause disassembly of the filament. In the absence of actin-binding proteins, the filament length is stable by a treadmilling mechanism (middle filament). Profilin enhances filament assembly by promoting ADP to ATP exchange on actin and by directing actin monomers to the barbed end of filaments (bottom filament). (Figure adapted from [8]). |
References
- Loisel TP., Boujemaa R., Pantaloni D. & Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 1999; 401(6753):613-6. [PMID: 10524632]
- Pollard TD., Blanchoin L. & Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 2000; 29:545-76. [PMID: 10940259]
- Wegner A. Head to tail polymerization of actin. J. Mol. Biol. 1976; 108(1):139-50. [PMID: 1003481]
- Carlier MF. & Pantaloni D. Control of actin dynamics in cell motility. J. Mol. Biol. 1997; 269(4):459-67. [PMID: 9217250]
- Alberts JB. & Odell GM. In silico reconstitution of Listeria propulsion exhibits nano-saltation. PLoS Biol. 2004; 2(12):e412. [PMID: 15562315]
- Rosenblatt J., Peluso P. & Mitchison TJ. The bulk of unpolymerized actin in Xenopus egg extracts is ATP-bound. Mol. Biol. Cell 1995; 6(2):227-36. [PMID: 7787248]
- Bugyi B. & Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys 2010; 39:449-70. [PMID: 20192778]
- Le Clainche C. & Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 2008; 88(2):489-513. [PMID: 18391171]