Functional Modules
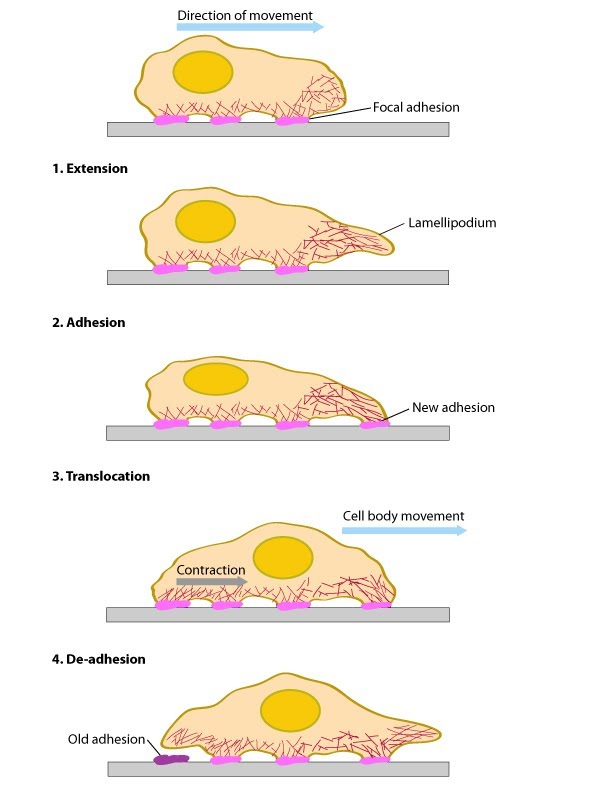
| 1.2 Steps in the Formation and Function of Lamella and LamellipodiaIn general, cell motility requires two types of forces: 1) a protruding force to extend the leading edge forward; and 2) traction forces to move the cell body [1] (reviewed in [2, 3]). These driving forces are functionally-integrated and they are modulated by actin filament dynamics [4, 5, 6, 7, 8, 9] (reviewed in [10, 11, 12]). Cell spreading and motility also require plasticity at the plasma membrane and cell adhesions (reviewed in [13] in addition to transcriptional control of gene expression [14]).The lamellipodia and lamella are two distinct regions of the cell that facilitate cell motility and function in mechanosensing mechanisms. These regions undergo defined steps in their formation and function as is described in this section. In summary, the steps of formation and function are:
The above mentioned activities produce net forward/backward cell movement or spreading of the plasma membrane in a specific direction (aka polarized movement). It should be noted however that the specific biophysical and biochemical parameters that are altered at each step have been difficult to measure due to the wide variety of motile structures that can be found in each cell at any given time. For example, migratory fibroblasts exhibit LP protrusion and retraction, ruffling, filopodial protrusion and retraction, bleb protrusion and retraction, trailing edge retraction, and quiescence in neighboring regions of the cell edge [5]. Fortunately, recent advances in technological systems that measure the temporal and spatial movement of single proteins in specific cell structures, whole cells, and tissues has greatly expanded our mechanistic understanding of cell motility and the composite functional modules [5, 8, 15, 16, 17](reviewed in [18]). Surprisingly, recent work has suggested that the basic mechanism for polarization and directional movement lies in the microtubules, which can be modified by their interaction with the actin-myosin system and cell-substrate adhesions [19]. Although there are numerous details that remain unresolved, it is abundantly clear that mechanical mechanisms are essential for coordinating the physical and biochemical processes that determine cell shape and locomotion. |
References
- Oliver T., Dembo M. & Jacobson K. Separation of propulsive and adhesive traction stresses in locomoting keratocytes. J. Cell Biol. 1999; 145(3):589-604. [PMID: 10225959]
- Elson EL., Felder SF., Jay PY., Kolodney MS. & Pasternak C. Forces in cell locomotion. Biochem. Soc. Symp. 1999; 65:299-314. [PMID: 10320946]
- Theriot JA. The polymerization motor. Traffic 2000; 1(1):19-28. [PMID: 11208055]
- Loisel TP., Boujemaa R., Pantaloni D. & Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 1999; 401(6753):613-6. [PMID: 10524632]
- Dubin-Thaler BJ., Hofman JM., Cai Y., Xenias H., Spielman I., Shneidman AV., David LA., Döbereiner HG., Wiggins CH. & Sheetz MP. Quantification of cell edge velocities and traction forces reveals distinct motility modules during cell spreading. PLoS ONE 2008; 3(11):e3735. [PMID: 19011687]
- Ponti A., Machacek M., Gupton SL., Waterman-Storer CM. & Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science 2004; 305(5691):1782-6. [PMID: 15375270]
- Gupton SL. & Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 2006; 125(7):1361-74. [PMID: 16814721]
- Giannone G., Dubin-Thaler BJ., Rossier O., Cai Y., Chaga O., Jiang G., Beaver W., Döbereiner HG., Freund Y., Borisy G. & Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell 2007; 128(3):561-75. [PMID: 17289574]
- Schaub S., Bohnet S., Laurent VM., Meister JJ. & Verkhovsky AB. Comparative maps of motion and assembly of filamentous actin and myosin II in migrating cells. Mol. Biol. Cell 2007; 18(10):3723-32. [PMID: 17634292]
- Carlier MF., Le Clainche C., Wiesner S. & Pantaloni D. Actin-based motility: from molecules to movement. Bioessays 2003; 25(4):336-45. [PMID: 12655641]
- Pollard TD. & Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003; 112(4):453-65. [PMID: 12600310]
- Le Clainche C. & Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 2008; 88(2):489-513. [PMID: 18391171]
- Lock JG., Wehrle-Haller B. & Strömblad S. Cell-matrix adhesion complexes: master control machinery of cell migration. Semin. Cancer Biol. 2008; 18(1):65-76. [PMID: 18023204]
- Dalby MJ., Riehle MO., Sutherland DS., Agheli H. & Curtis AS. Morphological and microarray analysis of human fibroblasts cultured on nanocolumns produced by colloidal lithography. Eur Cell Mater 2005; 9:1-8; discussion 8. [PMID: 15690263]
- Döbereiner HG., Dubin-Thaler BJ., Hofman JM., Xenias HS., Sims TN., Giannone G., Dustin ML., Wiggins CH. & Sheetz MP. Lateral membrane waves constitute a universal dynamic pattern of motile cells. Phys. Rev. Lett. 2006; 97(3):038102. [PMID: 16907546]
- Alexandrova AY., Arnold K., Schaub S., Vasiliev JM., Meister JJ., Bershadsky AD. & Verkhovsky AB. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS ONE 2008; 3(9):e3234. [PMID: 18800171]
- Tsukada Y., Aoki K., Nakamura T., Sakumura Y., Matsuda M. & Ishii S. Quantification of local morphodynamics and local GTPase activity by edge evolution tracking. PLoS Comput. Biol. 2008; 4(11):e1000223. [PMID: 19008941]
- Vogel V. & Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006; 7(4):265-75. [PMID: 16607289]
- Shutova MS., Alexandrova AY. & Vasiliev JM. Regulation of polarity in cells devoid of actin bundle system after treatment with inhibitors of myosin II activity. Cell Motil. Cytoskeleton 2008; 65(9):734-46. [PMID: 18615701]