How do actin monomers polymerize to form an actin filament?[Edit]
Actin filaments are highly dynamic and their polymerization is usually correlated to their disassembly. Generally actin filament polymerization occurs over three phases: A nucleation phase, an elongation phase and a steady state phase.
Step 3: Steady State Phase (Treadmilling)
 Figure 2. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the Cc
Figure 2. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the Cc
When the association rate of free ATP-G-actin is greater than the rate of subunit loss, the filament appears to grow, creating a ‘cap’ rich in ATP-subunits [10]. Conversely, when the association rate of free ATP-actin is lower than the rate of subunit loss, the filament is seen to shrink. When the association rate of free ATP-actin is equal to the rate of dissociation at the (-) end, no net growth occurs and this is known as ‘treadmilling’.
Table 2: Cellular concentrations (in µM) of key proteins in the actin system of diverse cells (if known, modified from [2] and [3]). * These values are representative of biological concentrations similar to those used for in vitro reconstitution experiments of bacterial motility and so are not cell type specific[3].
During the nucleation phase the formation of a stable ‘actin nucleus’ occurs. This is usually comprised of three actin monomers in complex. In the elongation phase monomers are rapidly added to the filament at the (+ve) or barbed end and this is often facilitated by additional elongation factors such as formin. In the steady state phase, the filament dynamics enter a state of equilibrium where monomer disassembly from the (-) end and polymerization at the (+) end is balanced and maintained by a critical concentration of monomers in the cytosol. This steady state assembly and disassembly is known as ‘treadmilling’.
Video: Actin filament assembly. The actin network is made up of filamentous actin (F-actin). These filaments are highly dynamic in nature and comprise monomers of G-actin bound to either ATP (yellow) or ADP (blue). Assembly is powered by ATP hydrolysis and filament nucleation happens spontaneously in vitro. Polymerization: Addition of ATP-actin occurs at the barbed end, leading to filament elongation. Elongation will continue whilst the rate of elongation is greater than the loss of ADP-actin from the pointed end. Profilin preferentially binds to ATP-actin, inhibits nucleation and accelerates filament elongation in vivo. Depolymerization: When the dissociation rate of ADP-actin exceeds the rate of ATP-actin association, the filament shrinks. In vivo, this is aided by cofilin, which can severe filaments into short fragments and promote subunit loss from the pointed ends. Actin treadmilling occurs when the rate of association of ATP-actin and the rate of loss of ADP-actin are balanced. [Video produced by MBInfo]
 Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.Step 1: Actin Nucleation
Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.Step 1: Actin Nucleation
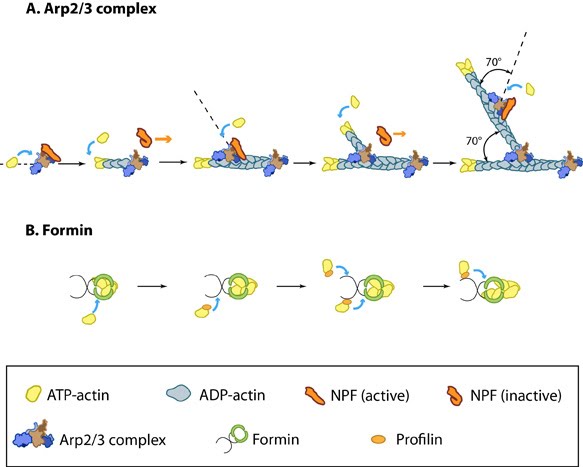 Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.
Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.The first step in actin polymerization is known as ‘nucleation’. This step sees the formation of an actin nucleus, which is essentially a complex of three actin monomers, from which an actin filament may elongate. Although actin monomers will spontaneously oligomerize in solution when present at a concentration above the critical concentration (Cc), these complexes are highly unstable. Actin nucleation therefore requires additional proteins known as ‘actin nucleators’ to promote the formation of a stable actin nucleus. These proteins include the Arp2/3 complex, formins, and the ‘tandem-monomer-binding nucleators’.
Step 2: Filament Elongation Phase
In the 2nd step, the rapid addition of actin monomers to the (+ve) end of the actin filament occurs. This process, known as elongation is mediated by proteins that translocate along the growing filament and simultaneously catalyze the addition of monomers to the filament end. Such proteins include members of the formin family of proteins[2]. For this process to occur, the (+) end of the filament must be exposed, and this means removal of capping protein.
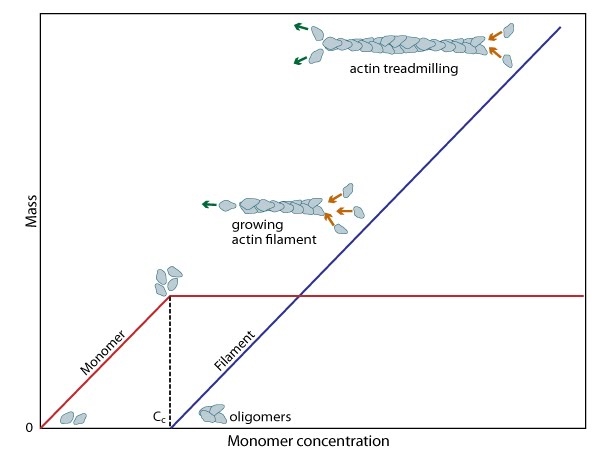 Figure 2. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the Cc
Figure 2. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the CcA steady pool of actin monomers must be maintained to enable a polymerization to continue beyond the rapid elongation phase. This is, in part, aided by the actin binding protein profilin, which promotes ADP to ATP nucleotide exchange on G-actin. However, the rate of monomer dissociation from the (-) end of the filament is also important. Dissociation of the subunits ultimately results from ATP hydrolysis, which induces a conformational change in the actin subunit that weakens its association with neighboring subunits (as reviewed in [8]). The concentration of actin monomers in the cytosol will either favor disassembly, or assembly of the actin filament,
and these values are known as the critical concentration (Cc). When the concentration of free subunits exceeds the Cc, filament elongation occurs spontaneously [9]. Importantly, the Cc usually varies between the filament (+) end and the (-) end. At the steady state, which is achieved when the rate of filament polymerization is equally balanced by filament disassembly, the free subunit concentration is higher than the Cc at the (+) end and lower than the Cc at the (-) end. This results in subunits being added to the (+) end and dissociating from the (-).
When the association rate of free ATP-G-actin is greater than the rate of subunit loss, the filament appears to grow, creating a ‘cap’ rich in ATP-subunits [10]. Conversely, when the association rate of free ATP-actin is lower than the rate of subunit loss, the filament is seen to shrink. When the association rate of free ATP-actin is equal to the rate of dissociation at the (-) end, no net growth occurs and this is known as ‘treadmilling’.
| Cc | Rate of G-actin addition | Rate of G-actin dissociation | |
| (-) end | 0.6 µM | 1.3 µM -1 s -1 | 0.8 s -1 |
| (+) end | 0.12 µM | 12 µM -1 s -1 | 1.4 s -1 |
| Protein | Acanthamoeba | Dictyostelium | Xenopusegg extract | S. cerevisiae | in silico Listeriamodel * |
| Polymerized actin (e.g. F-actin) | 100 | 90 | 4 | 2 | |
| Unpolymerized actin | 00 | 160 | 12 | 0.01 | 12 |
| Profilin | 100 | 5 | present | 5 |
|
| Thymosin-β4 | absent? | absent? |
20 | absent | |
| ADF/cofilin | 20 | <100 | 3 | present | 3 |
| Arp2/3 complex | 2-4 | present | present | 0.3 |
|
| Capping protein | 1 | 1 | 1 | 1 |
|
| Gelsolin | ? absent | ||||
| α-actinin | 4 | 3 | |||
| VASP | 0.5 |
||||
| Act A |
105/µm2 |