Dynamics and Components for Actin Filament Polymerization[Edit]
Actin filaments are dynamic structures whose growth and disassembly are tightly controlled by additional proteins. These proteins may either promote actin filament nucleation by stabilizing the actin nucleus, catalyze filament elongation or promote actin treadmilling. Some well established proteins that play such roles include the Arp2/3 complex, which facilitates nucleation of filament branches; profilin, which catalyzes ADP to ATP exchange and ADF/cofilin, which mediates filament disassembly. The cooperation between each component is extensive and each element has an optimal concentration.
 Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
Factors influencing actin filament length and treadmilling
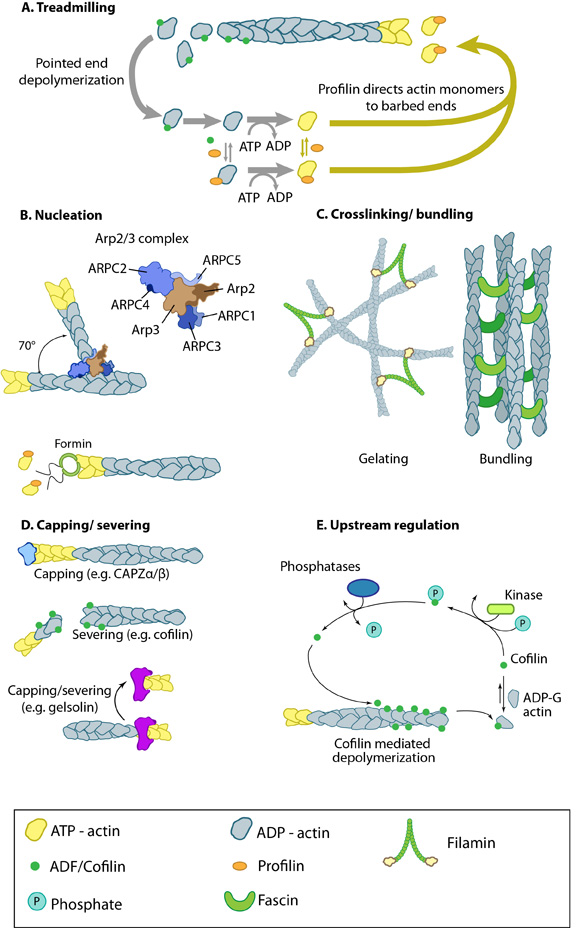 Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.