α-actinin[Edit]
α-actinin is an actin-binding protein [1] and component of the actin crosslinking functional modules; it lacks G-actin binding activity and lacks actin initiation/nucleation activity [2]. α-actinin is an important organizer of the cytoskeleton that belongs to the spectrin superfamily (which includes spectrin, dystrophin, and related homologues). α-actinin is present in a number of diverse organisms including protists, invertebrates, and birds; mammals have at least four α-actinin genes that together account for 6 different α-actinin proteins whose expression profile is tissue specific (reviewed in [3]).
All α-actinin proteins have a flexible amino-terminal F-actin binding domain (ABD, composed from two calponin homology [CH] domains), a central rod containing spectrin repeats (S or SR), and a carboxy-terminal calmodulin (CaM)-like domain composed of EF-hand calcium-binding motifs (see figure below). Calcium inhibits the association of non-muscle α-actinin isoforms with F-actin [4, 5, 6] whereas binding of the muscle isoforms is insensitive to calcium [7].
 Figure 1. Alpha (α)-actinin: This schematic diagram illustrates the molecular organization of α-actinin (reviewed in [3]) and provides examples for how α-actinin is represented in figures throughout this resource. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (actin binding domain (ABD) [8], β-integrin [9, 10], α-catenin [11] and vinculin [12, 13]).
Figure 1. Alpha (α)-actinin: This schematic diagram illustrates the molecular organization of α-actinin (reviewed in [3]) and provides examples for how α-actinin is represented in figures throughout this resource. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (actin binding domain (ABD) [8], β-integrin [9, 10], α-catenin [11] and vinculin [12, 13]).
The basic organization of the ABDs is quite similar in other members of the α-actinin superfamily such as filamin, fimbrin, and parvin [14]. α-actinin binds F-actin [8] and other molecules such as phophatidylinositol-bisphosphate (PIP2) [15], cell adhesion proteins (e.g. integrins [9, 10]) and signaling enzymes (e.g. PI3K [16]). It also interacts with vinculin through the 4th S repeat [13] and this interaction acts as a regulatory switch in adherens junctions [12]. Intramolecular contacts that sterically prevent α-actinin from interacting with actin filaments and integrins are relieved by PIP2 binding to the ABD [17] and this regulates α-actinin dynamics [18].
Dimerization of α-actinin via the rod domain is also essential for crosslinking actin [19] and for binding to other proteins (e.g. zyxin) [20], therefore, a dimer has functional domains at both ends [21]; this organization allows them to bind to adjacent actin filaments [22]. The smaller size of the α-actinin dimer combined with flexible hinges at the ABDs, makes α-actinin a versatile actin crosslinker capable of forming variable orientations and angles between actin filaments, as well as forming tighter bridges between filaments (such as those found in actin bundles) (reviewed in [3]).
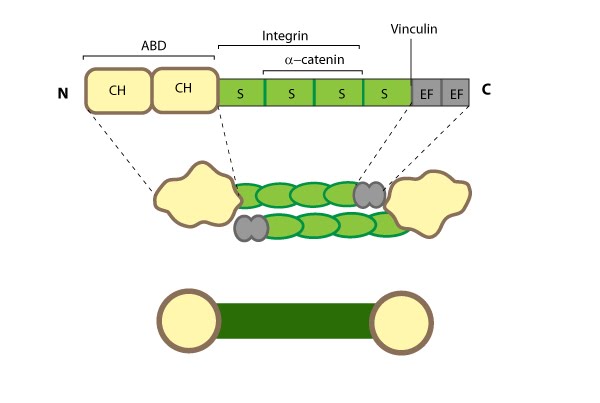 Figure 1. Alpha (α)-actinin: This schematic diagram illustrates the molecular organization of α-actinin (reviewed in [3]) and provides examples for how α-actinin is represented in figures throughout this resource. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (actin binding domain (ABD) [8], β-integrin [9, 10], α-catenin [11] and vinculin [12, 13]).
Figure 1. Alpha (α)-actinin: This schematic diagram illustrates the molecular organization of α-actinin (reviewed in [3]) and provides examples for how α-actinin is represented in figures throughout this resource. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (actin binding domain (ABD) [8], β-integrin [9, 10], α-catenin [11] and vinculin [12, 13]).The basic organization of the ABDs is quite similar in other members of the α-actinin superfamily such as filamin, fimbrin, and parvin [14]. α-actinin binds F-actin [8] and other molecules such as phophatidylinositol-bisphosphate (PIP2) [15], cell adhesion proteins (e.g. integrins [9, 10]) and signaling enzymes (e.g. PI3K [16]). It also interacts with vinculin through the 4th S repeat [13] and this interaction acts as a regulatory switch in adherens junctions [12]. Intramolecular contacts that sterically prevent α-actinin from interacting with actin filaments and integrins are relieved by PIP2 binding to the ABD [17] and this regulates α-actinin dynamics [18].
Dimerization of α-actinin via the rod domain is also essential for crosslinking actin [19] and for binding to other proteins (e.g. zyxin) [20], therefore, a dimer has functional domains at both ends [21]; this organization allows them to bind to adjacent actin filaments [22]. The smaller size of the α-actinin dimer combined with flexible hinges at the ABDs, makes α-actinin a versatile actin crosslinker capable of forming variable orientations and angles between actin filaments, as well as forming tighter bridges between filaments (such as those found in actin bundles) (reviewed in [3]).
α-actinin localization and function
α-actinin primarily influences the cohesiveness and mechanics of the cytoskeleton by cross-linking actin filaments and other cytoskeleton components to create a scaffold that imparts stability and forms a bridge between the cytoskeleton and signaling pathways. α-actinin interacts with numerous (~30) components in the cell (reviewed in [23]) and certain α-actinin isoforms (and related proteins) appear to be active in the nucleus (reviewed in [24]). α-actinin is mainly found at the leading edge of migrating cells and it is an important component of adhesion modules [25]. Dendritic spines are also rich in α-actinin and it appears to play a role in neuritic outgrowth [26]. Lastly, α-actinin is believed to be the primary crosslinking protein in stress fibers [27] and it plays a major role in the maturation of focal adhesions [28]. Localization of α-actinin to the plasma membrane is controlled by a number of interactions with membrane lipids and transmembrane receptors (reviewed in [2]). For example, binding of PIP2 at the plasma membrane causes a conformational change in the CaM-like domain that subsequently increases α-actinin’s affinity for actin and its ability to interact with other cytoskeletal components (e.g. titin [29]) (reviewed in [3]).