The Structure of Formin[Edit]
Actin filament assembly and disassembly are primary molecular processes that facilitate whole cell motility and the movement of subcellular structures. The initial stage in the formation of actin filaments is nucleation.
 Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domain
Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domain
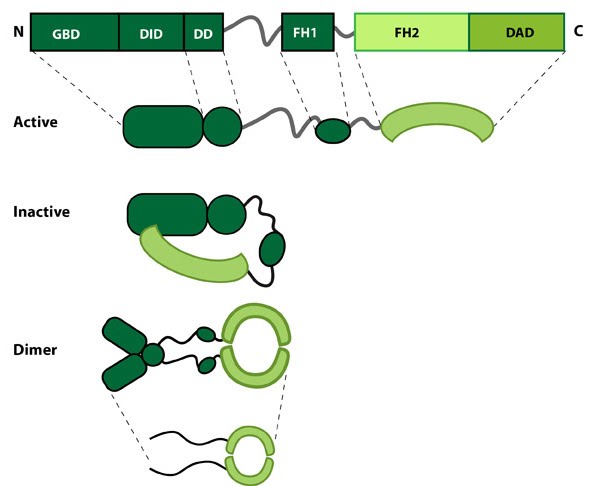 Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domain
Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domainNucleation is defined as the formation of a stable actin polymer from its monomeric units [5] and may also be facilitated by formin. Further to nucleation formins also facilitate the processive elongation of actin filaments, exclusively at the barbed end (reviewed in [6]). All members of this family share a common Formin homology 2 domain (FH2) and all but one (Dictyostelium discoideum formin, ForC) feature a FH1 domain. The diaphanous auto-regulatory domain (DAD), diaphanous inhibitory domain (DID) and the GTPase binding domain are also commonly identified in members of the formin family, though not found in all members. Each formin functions as a homodimer that shares structural similarity to F-actin. Each of these domains are functionally important, playing crucial roles in either the translocation of the protein along the growing actin filament, addition of actin monomers to the actin filament or in the regulation of the protein.