Actin Binding
Actin Binding Proteins[Edit]
Many proteins and small molecule ligands bind to actin. In doing so they modulate the dynamics of the actin cytoskeleton, catalyze actin filament assembly or promote actin filament disassembly. This is not only integral to broader cellular processes such as cell migration and mechanosensing, but may also be exploited for experimental purposes. Proteins that bind to actin filaments direct the location, rate, and timing of actin filament assembly and disassembly. There is great structural diversity in the types of proteins which bind to actin, but the actin binding domains (ABD) themselves can be grouped according to the conserved structures they form (reviewed in [1]).
Examples of common actin binding domains include:
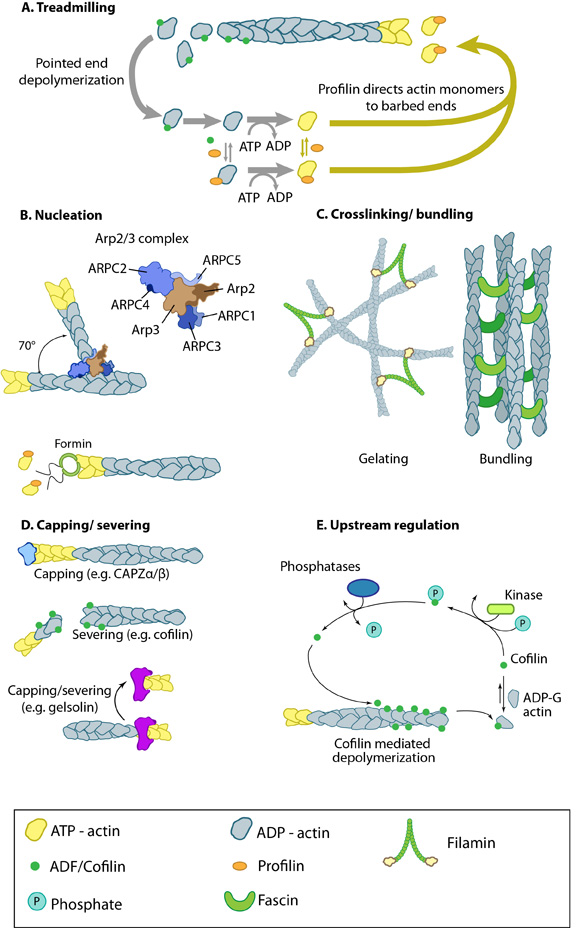 Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
An example of how actin binding proteins may alter the dynamics and structure of the actin filament network is seen in the actin binding protein fascin which produces actin filament bundles during filopodia formation. Importantly, different types of crosslinking proteins will give rise to different types of filament based structures and will also modulate physical dynamics of the network to varying degrees (reviewed in [2, 3, 4]).
Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
An example of how actin binding proteins may alter the dynamics and structure of the actin filament network is seen in the actin binding protein fascin which produces actin filament bundles during filopodia formation. Importantly, different types of crosslinking proteins will give rise to different types of filament based structures and will also modulate physical dynamics of the network to varying degrees (reviewed in [2, 3, 4]).
Although functionally similar, other cross-linking proteins may promote the formation of larger networks, or filamentous structures, in different regions of the cell. Generally, proteins with similar functions (e.g. fascin, α-actinin) act cooperatively to enhance the mechanical integrity and responsiveness of the network [5] (reviewed in [6]).
- calponin-homology (CH) domain
- formin-homology-2 (FH2) domain
- WASp-homology-2 (WH2) domain
- actin-depolymerizing factor/cofilin (ADF/cofilin) domain
- gelsolin-homology domain
- myosin motor domain
 Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
Figure 1. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.Although functionally similar, other cross-linking proteins may promote the formation of larger networks, or filamentous structures, in different regions of the cell. Generally, proteins with similar functions (e.g. fascin, α-actinin) act cooperatively to enhance the mechanical integrity and responsiveness of the network [5] (reviewed in [6]).
Small Molecules/Drugs that Bind to Actin[Edit]
Although actin-binding drugs have proven ineffective as pharmacological treatments due to poor uptake into cells or specific adverse effects, they are still widely used in research for their ability to alter actin filament dynamics. Toxins such as phalloidins, cytochalasins, latrunculin A, and jasplakinolide are naturally occurring small molecules that bind to actin and alter its polymerization.
As experimental agents, the phalloidins are limited by their inability to permeate the cellular membrane of living cells. Despite this their effect on polymerization differs from other commonly used agents and therefore the phalloidins are valuable agents for in vitro experiments.
Phalloidins bind to actin filaments at a ratio of one molecule for either one or two actin protomers and essentially lock adjacent actin subunits together. This shifts the equilibrium to favour filament formation over filament disassembly. As filament formation is normally a balance between monomer association and filament dissociation, blocking the latter step reduces the critical concentration of free monomers required to maintain filament elongation. (reviewed in [7]).
Phalloidins bind to actin filaments at a ratio of one molecule for either one or two actin protomers and essentially lock adjacent actin subunits together. This shifts the equilibrium to favour filament formation over filament disassembly. As filament formation is normally a balance between monomer association and filament dissociation, blocking the latter step reduces the critical concentration of free monomers required to maintain filament elongation. (reviewed in [7]).
Latrunculin A
Latrunculin A is a natural toxin purified from the red sea sponge Latrunculia magnifica. It was initially found to inhibit actin polymerization and disrupt the arrangement of F-actin within non-muscle cells. Further studies identified a 1:1 binding ratio between Latrunculin A and G-actin monomers [8]. Through the sequestration of actin monomers, polymerization is prevented and thus filament disassembly is promoted [9]. Studies investigating the binding mechanism of Latrunculin A to actin have revealed its presence does not inhibit profilin binding, suggesting the drug binds at a different site. It has been postulated that the drug binds at the ATP binding site, as it was shown to inhibit nucleotide exchange on actin subunits [10]. Binding of thymosin β4 to actin however is also inhibited by Latrunculin A, despite its binding site lying more than 20Å from the nucleotide binding site. This suggests either an allosteric effect on the nucleotide binding site, or the thymosin binding site, with the actual binding site remaining unclear [10].
Cytochalasin D
Cytochalasin D is the most commonly used drug from the cytochalasin class of small molecules. Various effects were reported from this class of compounds, including inhibition of monosaccaride transport across the cellular membrane [11], however its effect on actin polymerization is the most prominent.
Unlike Latrunculin A, the cytochalasins bind actin filaments rather than G-actin monomers. Binding occurs at a ratio of one molecule per filament, and binding occurs selectively at the barbed end of the filaments. This effectively blocks association (and dissociation) of actin monomers at the barbed end, halting further elongation of filaments [7].
Unlike Latrunculin A, the cytochalasins bind actin filaments rather than G-actin monomers. Binding occurs at a ratio of one molecule per filament, and binding occurs selectively at the barbed end of the filaments. This effectively blocks association (and dissociation) of actin monomers at the barbed end, halting further elongation of filaments [7].