Extra-cellular matrix disassembly
Content
Matrix metalloproteinases (MMPs) facilitate extracellular matrix disassembly[Edit]
Extracellular matrix disassembly is the degradation of the matrix components by proteases such as matrix metalloproteinases, and is a key step in normal physiological functions such as embryonic development and tissue remodeling, as well as in pathological conditions such as cancer progression.
 Figure 1. Matrix metalloproteinases (MMPs): This schematic diagram illustrates the molecular organization of MMPs (reviewed in [1]). Relevant domains believed to be important for MMP activity are highlighted (reviewed in [1]).
Mammalian MMPs comprise three major domains; an N terminal auto-inhibitory pro-domain, a catalytic domain and a C terminal hemopexin domain (see Figure at top right) [3]. MMPs are produced as inactive precursors. Their activity is inhibited through a conserved cysteine residue residing within the N terminal auto-inhibitory pro-domain. This residue interacts with the zinc ion bound to the catalytic domain and prevents cleavage of the substrate. The catalytic domain is connected to the hemopexin domain via a flexible linker peptide. The hemopexin domain forms a beta propeller structure, made of four ‘blades’ each of which represent a single hemopexin module. This domain regulates protein-protein interactions, substrate specificity and enzyme activity via interactions with tissue inhibitors of matrix metalloproteinases (TIMPs) [4].
Figure 1. Matrix metalloproteinases (MMPs): This schematic diagram illustrates the molecular organization of MMPs (reviewed in [1]). Relevant domains believed to be important for MMP activity are highlighted (reviewed in [1]).
Mammalian MMPs comprise three major domains; an N terminal auto-inhibitory pro-domain, a catalytic domain and a C terminal hemopexin domain (see Figure at top right) [3]. MMPs are produced as inactive precursors. Their activity is inhibited through a conserved cysteine residue residing within the N terminal auto-inhibitory pro-domain. This residue interacts with the zinc ion bound to the catalytic domain and prevents cleavage of the substrate. The catalytic domain is connected to the hemopexin domain via a flexible linker peptide. The hemopexin domain forms a beta propeller structure, made of four ‘blades’ each of which represent a single hemopexin module. This domain regulates protein-protein interactions, substrate specificity and enzyme activity via interactions with tissue inhibitors of matrix metalloproteinases (TIMPs) [4].
Variations in MMP structure exist (see Figure: Variations in MMP structure at bottom), with the addition or loss of certain domains (as reviewed in [3]). For example; MMP7 and MMP26 lack the hemopexin domain. MMP2 and MMP9 have additional fibronectin domain inserts within the catalytic domain. Membrane bound MMPs have additional membrane linkage domains, such as a GPI (glycosylphosphatidylinositol) anchor in the case of MMPs 14, 15, 16 and 24.
Matrix metalloproteinases (MMPs)
Matrix metalloproteinases (MMPs) are a family of proteases that digest components of the extracellular matrix (ECM) and surface receptors. The first MMP was identified in the 1960s as an enzyme which could digest the ECM component collagen and so was named collagenase [1]. Following this initial discovery a whole family of structurally related proteases were discovered. These proteases were all able to digest ECM components and did so in a zinc dependent manner. This led to the renaming of this protein family to matrix metalloproteinases. There are currently 23 human MMPs and 24 mouse MMPs [2], in addition to MMP homologs in invertebrates such as the fruit fly Drosophila melanogaster [2].MMP Structure
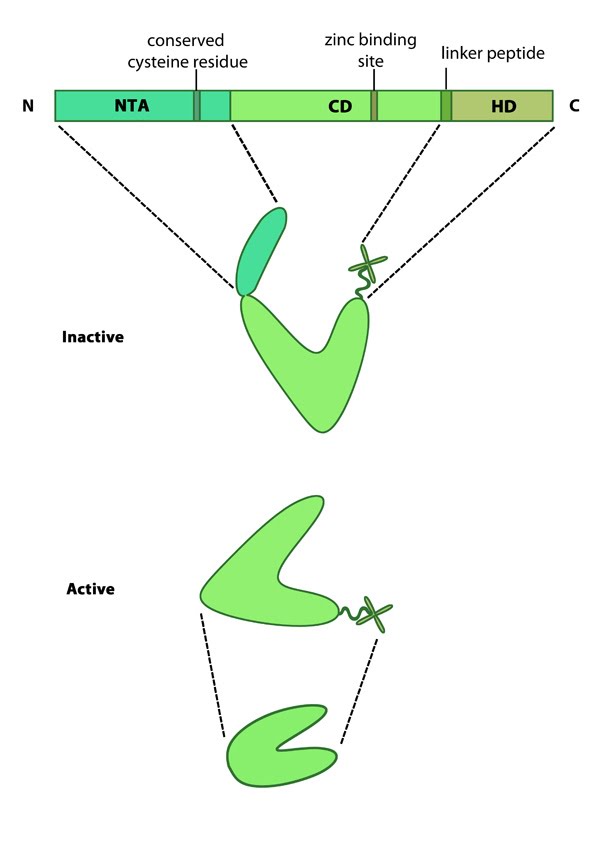 Figure 1. Matrix metalloproteinases (MMPs): This schematic diagram illustrates the molecular organization of MMPs (reviewed in [1]). Relevant domains believed to be important for MMP activity are highlighted (reviewed in [1]).
Figure 1. Matrix metalloproteinases (MMPs): This schematic diagram illustrates the molecular organization of MMPs (reviewed in [1]). Relevant domains believed to be important for MMP activity are highlighted (reviewed in [1]).Variations in MMP structure exist (see Figure: Variations in MMP structure at bottom), with the addition or loss of certain domains (as reviewed in [3]). For example; MMP7 and MMP26 lack the hemopexin domain. MMP2 and MMP9 have additional fibronectin domain inserts within the catalytic domain. Membrane bound MMPs have additional membrane linkage domains, such as a GPI (glycosylphosphatidylinositol) anchor in the case of MMPs 14, 15, 16 and 24.
Activation of MMPs[Edit]
MMPs are produced as inactive precursors which are exposed to the extracellular milieu either via secretion or translocation to the membrane. Once exposed to the extracellular environment they are activated through proteolytic cleavage of the N terminal autoinhibitory domain.  Figure 2. Variations in MMP structure: This schematic diagram illustrates variations on the common MMP structure (reviewed in [1]). (A) MMP2 and MMP9 have additional fibronectin domain inserts within the catalytic domain (B)
Membrane bound MMPs (MT-MMPs) contain an additional membrane linkage
domain attached to the hemopexin domain, in the form of either a GPI
(glycosylphosphatidylinositol) anchor or a transmembrane domain. (C) Minimal MMPs, such as MMP7 and MMP26, do not contain a linker peptide or hemopexin domain.Cleavage is carried out by a range of different proteases including the MMPs themselves, as well as tissue proteases such as tryptase, plasmin and kallikrein [4]. Following activation, MMPs are able to digest various ECM components including a host of collagen types (I-V, VII, X, XI), gelatin, laminin, basement membrane proteins entactin and perlecan and ECM glycoproteins such as tenascin [4]. Active MMPs are involved in a wide range of cellular processes including; tissue remodeling, cell survival, immune cell migration and cancer pathology [3].
Figure 2. Variations in MMP structure: This schematic diagram illustrates variations on the common MMP structure (reviewed in [1]). (A) MMP2 and MMP9 have additional fibronectin domain inserts within the catalytic domain (B)
Membrane bound MMPs (MT-MMPs) contain an additional membrane linkage
domain attached to the hemopexin domain, in the form of either a GPI
(glycosylphosphatidylinositol) anchor or a transmembrane domain. (C) Minimal MMPs, such as MMP7 and MMP26, do not contain a linker peptide or hemopexin domain.Cleavage is carried out by a range of different proteases including the MMPs themselves, as well as tissue proteases such as tryptase, plasmin and kallikrein [4]. Following activation, MMPs are able to digest various ECM components including a host of collagen types (I-V, VII, X, XI), gelatin, laminin, basement membrane proteins entactin and perlecan and ECM glycoproteins such as tenascin [4]. Active MMPs are involved in a wide range of cellular processes including; tissue remodeling, cell survival, immune cell migration and cancer pathology [3].
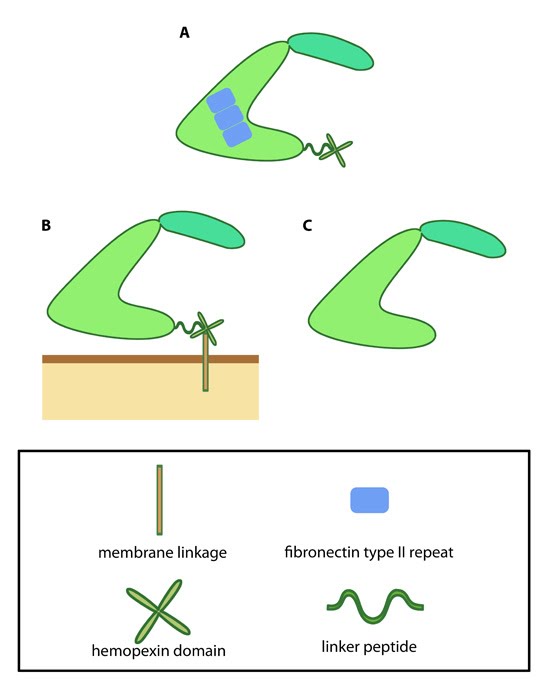 Figure 2. Variations in MMP structure: This schematic diagram illustrates variations on the common MMP structure (reviewed in [1]). (A) MMP2 and MMP9 have additional fibronectin domain inserts within the catalytic domain (B)
Membrane bound MMPs (MT-MMPs) contain an additional membrane linkage
domain attached to the hemopexin domain, in the form of either a GPI
(glycosylphosphatidylinositol) anchor or a transmembrane domain. (C) Minimal MMPs, such as MMP7 and MMP26, do not contain a linker peptide or hemopexin domain.
Figure 2. Variations in MMP structure: This schematic diagram illustrates variations on the common MMP structure (reviewed in [1]). (A) MMP2 and MMP9 have additional fibronectin domain inserts within the catalytic domain (B)
Membrane bound MMPs (MT-MMPs) contain an additional membrane linkage
domain attached to the hemopexin domain, in the form of either a GPI
(glycosylphosphatidylinositol) anchor or a transmembrane domain. (C) Minimal MMPs, such as MMP7 and MMP26, do not contain a linker peptide or hemopexin domain.