Integrin-Mediated Signalling Pathway
Content
Introduction to integrin and its structure[Edit]
Integrins are proteins that function mechanically, by attaching the cell cytoskeleton to the extracellular matrix (ECM), and biochemically, by sensing whether adhesion has occurred. The integrin family of proteins consists of alpha and beta subtypes, which form transmembrane heterodimers. Integrins function as adhesion receptors for extracellular ligands and transduce biochemical signals into the cell, through downstream effector proteins. Remarkably, they function bidirectionally, meaning they can transmit information both outside-in and inside-out (reviewed in [1, 2]).
 Figure 1. Integrin_alpha_chain: Integrin α subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin α subunit. The αI domain is present in some subtypes of the α subunit.
Each integrin heterodimer consists of an alpha (α) and a beta (β) subunit associated by noncovalent interactions forming an extracellular ligand-binding head, two multi-domain `legs’, two single-pass transmembrane helices and two short cytoplasmic tails. The α and β groups show no homology to each other,however, conserved regions are found among subtypes of both groups.
Figure 1. Integrin_alpha_chain: Integrin α subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin α subunit. The αI domain is present in some subtypes of the α subunit.
Each integrin heterodimer consists of an alpha (α) and a beta (β) subunit associated by noncovalent interactions forming an extracellular ligand-binding head, two multi-domain `legs’, two single-pass transmembrane helices and two short cytoplasmic tails. The α and β groups show no homology to each other,however, conserved regions are found among subtypes of both groups.
The α subunit leg consists of a thigh and 2 calf domains that support the ligand binding head formed by a β-propeller domain with 7 repeats forming the blades (shown as a cylinder in the figure below). Some of the propeller blade domains contain calcium binding EF-hand domains on the lower side; these allosterically affect ligand binding [3]. An additional αI (interactive) domain containing ~200 residues is present in some vertebrate α chains [4] (nine human α subtypes) between the propeller repeats 2 and 3 [5]. This contains a metal-ion dependent adhesion site (MIDAS) which is important for ligand binding.
 Figure 2. Integrin_beta_chain: Integrin β subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin β subunit.
The β subunit comprises of 4 cysteine-rich epidermal growth factor (EGF) repeats, a hybrid domain (split in sequence), an I-like domain (βI) and a plexin-sempahorin-integrin (PSI) domain. Similar to αI, the contains βI domain contains a MIDAS site for ligand binding and additional regulatory site “adjacent to MIDAS” or ADMIDAS, inhibited by Ca2+ and activated by Mn2+ [3] for ligand binding.
Figure 2. Integrin_beta_chain: Integrin β subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin β subunit.
The β subunit comprises of 4 cysteine-rich epidermal growth factor (EGF) repeats, a hybrid domain (split in sequence), an I-like domain (βI) and a plexin-sempahorin-integrin (PSI) domain. Similar to αI, the contains βI domain contains a MIDAS site for ligand binding and additional regulatory site “adjacent to MIDAS” or ADMIDAS, inhibited by Ca2+ and activated by Mn2+ [3] for ligand binding.
i) Ligand binding
The βI domain binds ligand together with the β-propeller or with αI (if present) through MIDAS in a Mg2+ dependent fashion [6]at the interface in the headpiece. While Asp carboxyl group coordinates the βI MIDAS ion Mg2+, side chain hydrogen of the Arg of the RGD ligand binds directly to the Asp in domains 2 and 3 of β-propeller [7].
 Figure 3. Integrin dimer structure: Globular domain structures of α and β subunits in a stable dimer. Ligand binding happens at the interface of the αI (left panel) or β-propeller (right panel) and the βI domain.
ii) Dimerization
Figure 3. Integrin dimer structure: Globular domain structures of α and β subunits in a stable dimer. Ligand binding happens at the interface of the αI (left panel) or β-propeller (right panel) and the βI domain.
ii) Dimerization
Dimerization occurs via the β-propeller surface on the α chain and the hybrid domain in the β chain in the cytoplasm [8]. The sequences at these interacting surfaces seem to control the specificity of chain selection. The dimers have been shown to be stabilized and remain inactive by hydrophobic interactions and electrostatic salt bridges at the outer- and inner-membrane proximal regions respectively [9, 10].
iii) Interactions
The cytoplasmic tail of β-chain is known to bind to protein adaptors through NPxY/F motifs [11]; this activates the integrins by breaking the salt bridge between the dimer (reviewed in [12, 13]). In general, the adaptor proteins promote linkage to actin [14], however intermediate filaments have also been implicated via vimentin [15, 16].
Scaffolding adaptors (e.g. paxillin, kindlin) forms bridges between focal adhesion proteins
Catalytic adaptors (e.g. focal adhesion kinase, integrin-linked kinase, Src) propagate signal transduction from adhesion sitesInteractions via α-tail are not well established due to sequence variability, however, α-tail is implicated in the cell-type specific integrin activation through binding proteins that modulate downstream signaling [17, 18]. Phosphorylation state of cytoplasmic tail residues modulate the competition between adaptors for binding and hence the subsequent cytoskeletal interactions of integrins and response (reviewed in [19]).
The role of protein structure in ligand affinity modulation, signaling and dynamics of surface distribution of integrins is reviewed in[20].
Protein Structure
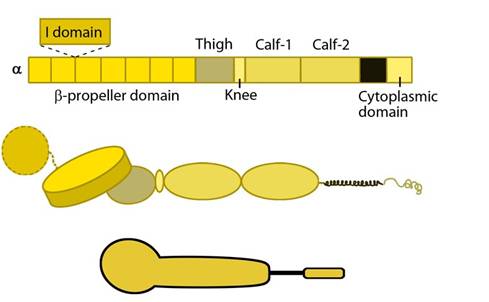 Figure 1. Integrin_alpha_chain: Integrin α subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin α subunit. The αI domain is present in some subtypes of the α subunit.
Figure 1. Integrin_alpha_chain: Integrin α subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin α subunit. The αI domain is present in some subtypes of the α subunit.The α subunit leg consists of a thigh and 2 calf domains that support the ligand binding head formed by a β-propeller domain with 7 repeats forming the blades (shown as a cylinder in the figure below). Some of the propeller blade domains contain calcium binding EF-hand domains on the lower side; these allosterically affect ligand binding [3]. An additional αI (interactive) domain containing ~200 residues is present in some vertebrate α chains [4] (nine human α subtypes) between the propeller repeats 2 and 3 [5]. This contains a metal-ion dependent adhesion site (MIDAS) which is important for ligand binding.
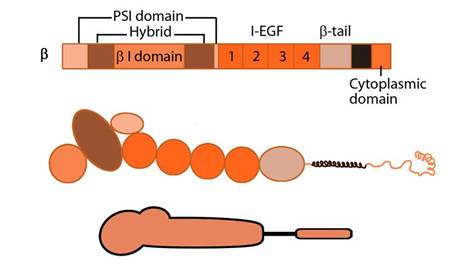 Figure 2. Integrin_beta_chain: Integrin β subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin β subunit.
Figure 2. Integrin_beta_chain: Integrin β subunit domains: Top: Linear domain arrangement. Middle: The globular structure formed by protein domains. Bottom: simplified version of the integrin β subunit.i) Ligand binding
The βI domain binds ligand together with the β-propeller or with αI (if present) through MIDAS in a Mg2+ dependent fashion [6]at the interface in the headpiece. While Asp carboxyl group coordinates the βI MIDAS ion Mg2+, side chain hydrogen of the Arg of the RGD ligand binds directly to the Asp in domains 2 and 3 of β-propeller [7].
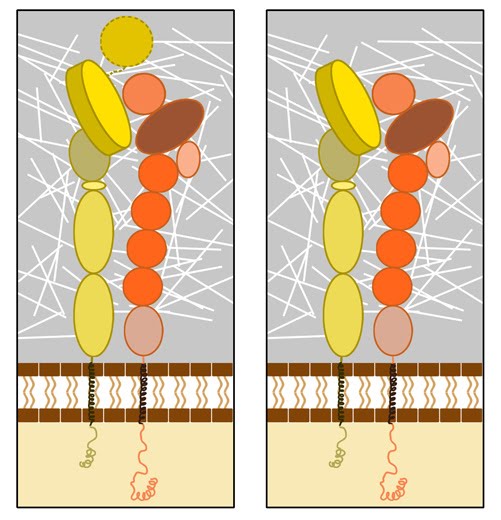 Figure 3. Integrin dimer structure: Globular domain structures of α and β subunits in a stable dimer. Ligand binding happens at the interface of the αI (left panel) or β-propeller (right panel) and the βI domain.
Figure 3. Integrin dimer structure: Globular domain structures of α and β subunits in a stable dimer. Ligand binding happens at the interface of the αI (left panel) or β-propeller (right panel) and the βI domain.Dimerization occurs via the β-propeller surface on the α chain and the hybrid domain in the β chain in the cytoplasm [8]. The sequences at these interacting surfaces seem to control the specificity of chain selection. The dimers have been shown to be stabilized and remain inactive by hydrophobic interactions and electrostatic salt bridges at the outer- and inner-membrane proximal regions respectively [9, 10].
iii) Interactions
The cytoplasmic tail of β-chain is known to bind to protein adaptors through NPxY/F motifs [11]; this activates the integrins by breaking the salt bridge between the dimer (reviewed in [12, 13]). In general, the adaptor proteins promote linkage to actin [14], however intermediate filaments have also been implicated via vimentin [15, 16].
Protein adaptors that bind to integrin cytoplasmic tails:
Structural adaptors (e.g. talin, filamin, tensin) link integrins directly to the cytoskeletonScaffolding adaptors (e.g. paxillin, kindlin) forms bridges between focal adhesion proteins
Catalytic adaptors (e.g. focal adhesion kinase, integrin-linked kinase, Src) propagate signal transduction from adhesion sitesInteractions via α-tail are not well established due to sequence variability, however, α-tail is implicated in the cell-type specific integrin activation through binding proteins that modulate downstream signaling [17, 18]. Phosphorylation state of cytoplasmic tail residues modulate the competition between adaptors for binding and hence the subsequent cytoskeletal interactions of integrins and response (reviewed in [19]).
The role of protein structure in ligand affinity modulation, signaling and dynamics of surface distribution of integrins is reviewed in[20].
Integrin-Ligand specificity[Edit]
Humans have at least 18 α subtypes and 8 β subtypes which together generate 24 known binding pairs for the integrin heterodimer (reviewed in [12, 21]). The α subunit of the integrin heterodimer especially the αI domain determines the ligand specificity for cell-ECM adhesion (reviewed in [22]). The characteristic of integrin subunits is their ability to bind diverse matrix molecules imparted by the heterogeneity of the monomers; this plasticity is instrumental for cell-ECM binding and subsequent mechanotransduction events.
The amino acid sequence: arginine-glycine-aspartic acid, or RGD motif, is commonly accepted as a general integrin-binding motif on target ligands, however, individual integrins can also recognize other protein-specific motifs (reviewed in [12, 21, 22]).
Common ECM components that are bound by integrins (with respective recognition sequence) are
* Fibronectin (RGD, LDV)
* Collagen (triple helical GFOGER)
* Laminin
* Vitronectin (RGD)
* Fibrinogen(RGD)
* Thrombospondin
* Glycoproteins (e.g. tenascin C, osteopontin, nefronectin)
Immunologically important integrin ligands are the inter-cellular adhesion molecules (ICAMs), immunoglobulin superfamily members present on inflamed endothelium and antigen-presenting cells.
Integrins are broadly grouped into four categories based on their ligand-specificity (reviewed in [21]):
RGD receptors (α5β1, αVβ3, αVβ1,αVβ5, αVβ6, αVβ8, and αIIbβ3)
Laminin receptors (α1β1, α2β1, α3β1, α6β1, α7β1, and α6β4)
Leukocyte-specific receptors (αLβ2, αMβ2, αXβ2, and αDβ2)
Collagen receptors (α1β1, α2β1, α3β1, α10β1, and α11β1)
The amino acid sequence: arginine-glycine-aspartic acid, or RGD motif, is commonly accepted as a general integrin-binding motif on target ligands, however, individual integrins can also recognize other protein-specific motifs (reviewed in [12, 21, 22]).
Common ECM components that are bound by integrins (with respective recognition sequence) are
* Fibronectin (RGD, LDV)
* Collagen (triple helical GFOGER)
* Laminin
* Vitronectin (RGD)
* Fibrinogen(RGD)
* Thrombospondin
* Glycoproteins (e.g. tenascin C, osteopontin, nefronectin)
Immunologically important integrin ligands are the inter-cellular adhesion molecules (ICAMs), immunoglobulin superfamily members present on inflamed endothelium and antigen-presenting cells.
Integrins are broadly grouped into four categories based on their ligand-specificity (reviewed in [21]):
RGD receptors (α5β1, αVβ3, αVβ1,αVβ5, αVβ6, αVβ8, and αIIbβ3)
Laminin receptors (α1β1, α2β1, α3β1, α6β1, α7β1, and α6β4)
Leukocyte-specific receptors (αLβ2, αMβ2, αXβ2, and αDβ2)
Collagen receptors (α1β1, α2β1, α3β1, α10β1, and α11β1)
Localization of Integrin[Edit]
The concentration of β subunits is generally high while that of α subunit is the factor limiting the amount of heterodimers that localize on the membrane [23]. Liganded integrins diffuse along the membrane and are known to preferentially couple actin at the leading edge [24], cluster, get pulled backward as the cell spreads or moves forward and subsequently cycled during motility [25,26].
Integrins are found in specialized cell-cell adhesions and in most cell-matrix adhesions of static cells as well as motility structures such as filopodia,lamellipodia and podosomes. While the β1 subfamily is commonly found on a wide variety of cells, some integrins are limited to certain cell types or tissues [12] as listed in the table.
Integrins are found in specialized cell-cell adhesions and in most cell-matrix adhesions of static cells as well as motility structures such as filopodia,lamellipodia and podosomes. While the β1 subfamily is commonly found on a wide variety of cells, some integrins are limited to certain cell types or tissues [12] as listed in the table.
Table: Cell-/tissue-specific localization of integrin subtypes
| Integrin subtypes | Cell/ tissue of localization |
| αIIbβ3 |
Platelets |
| α6β4 | Keratinocytes |
| αEβ7 | T-cells, dendritic cells and mast cells of mucosal tissue |
| α4β1, β2 integrins |
Leukocytes |
| α4β7 | Memory T-cells |
Integrin clustering[Edit]
Clustering happens by integrin diffusion, multivalent ligand binding leading to TM homodimerization or inside-out signals. For example, in lymphocytes Rap1 GTPase and its effector RAPL (regulator for cell adhesion and polarization enriched in lymphoid tissues) have been shown to regulate patchy distribution of integrin LFA-1 [27]. Nanolithographic study provides strong evidence that controlled spatial organization of liganded integrins in nanoclusters is essential for effective signaling and is independent of global density [28]. Also, the minimum cluster area required for stable adhesion formation and force transduction is determined by the adhesive force, cytoskeletal tension and the force-transmitting structural linkage. Thus it is not a constant and has a dynamic threshold [29].
Nanoclustering is driven by the interaction of N-terminal of vinculin with talin [30], which in turn is promoted by contractile forces. Some talin-integrin interactions are also essential to prevent re-association of the separated integrin tails and maintain the activated integrins in a clustering-competent form [31]. Besides increasing the avidity (valency) of ligand binding and clustering contributes to adhesion strength and outside-in signaling [25, 32]. However, whether clustering triggers outside-in signaling to facilitate integrin activation or happens after integrin activation is uncertain (reviewed in [33]).
Integrin functions[Edit]
For integrin to function as a bidirectional signal transmitter,
 Figure 4. Cellular responses elicited by integrin signaling: In response to physical/chemical properties of the matrix and growth factors in the environment (outside-in signaling), integrins bind ligands and get activated. Accordingly, a variety of signaling pathways can be triggered mainly through the different kinases as mentioned above. These can bring about changes in one or more cellular events (short term responses) that eventually result in global (long term) responses in cellular behavior. Adapted from [34].1) Integrins undergo a process called activation, during which, conformational changes expose the headpiece (βI and hybrid domain) for ligand binding [35, 36, 37, 38]. This can be initiated by binding of adaptor proteins and/or ligands.
Figure 4. Cellular responses elicited by integrin signaling: In response to physical/chemical properties of the matrix and growth factors in the environment (outside-in signaling), integrins bind ligands and get activated. Accordingly, a variety of signaling pathways can be triggered mainly through the different kinases as mentioned above. These can bring about changes in one or more cellular events (short term responses) that eventually result in global (long term) responses in cellular behavior. Adapted from [34].1) Integrins undergo a process called activation, during which, conformational changes expose the headpiece (βI and hybrid domain) for ligand binding [35, 36, 37, 38]. This can be initiated by binding of adaptor proteins and/or ligands.
2) They connect to the cytoskeleton through adaptor proteins binding to the integrin cytoplasmic domains.
3) Integrins microcluster laterally for efficient ligand binding.
Upon activation, integrins are capable of triggering a variety of signal transduction cascades based on the context, ligand (matrix components/growth factors) and substrate stiffness that result in a variety of short-term and long-term responses [37].
Involvement of the different combination of α and β subtypes in a variety of in vivo functions including role in cell behavior and tissue organization have been demonstrated by knockout mice studies (reviewed in [39, 40, 41, 42]; See table below).
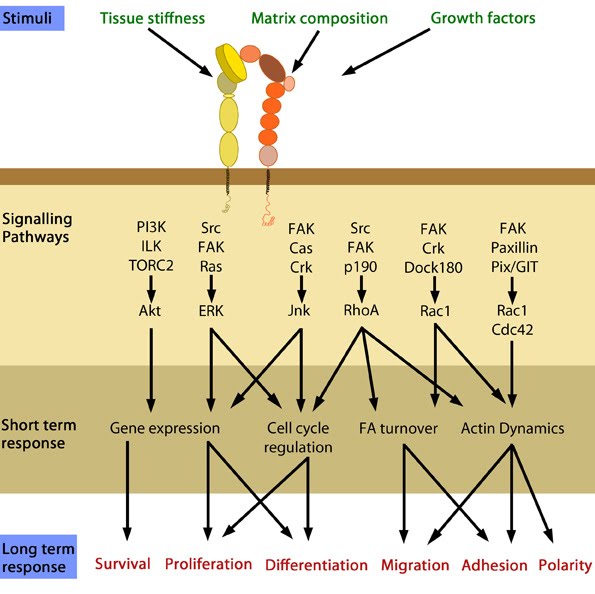 Figure 4. Cellular responses elicited by integrin signaling: In response to physical/chemical properties of the matrix and growth factors in the environment (outside-in signaling), integrins bind ligands and get activated. Accordingly, a variety of signaling pathways can be triggered mainly through the different kinases as mentioned above. These can bring about changes in one or more cellular events (short term responses) that eventually result in global (long term) responses in cellular behavior. Adapted from [34].
Figure 4. Cellular responses elicited by integrin signaling: In response to physical/chemical properties of the matrix and growth factors in the environment (outside-in signaling), integrins bind ligands and get activated. Accordingly, a variety of signaling pathways can be triggered mainly through the different kinases as mentioned above. These can bring about changes in one or more cellular events (short term responses) that eventually result in global (long term) responses in cellular behavior. Adapted from [34].2) They connect to the cytoskeleton through adaptor proteins binding to the integrin cytoplasmic domains.
3) Integrins microcluster laterally for efficient ligand binding.
Upon activation, integrins are capable of triggering a variety of signal transduction cascades based on the context, ligand (matrix components/growth factors) and substrate stiffness that result in a variety of short-term and long-term responses [37].
Involvement of the different combination of α and β subtypes in a variety of in vivo functions including role in cell behavior and tissue organization have been demonstrated by knockout mice studies (reviewed in [39, 40, 41, 42]; See table below).
Table: Role of some integrin subtypes in specific in vivo functions
| Integrin type |
In vivo function |
| β1 integrins |
Development |
| αV |
Vasculogenesis |
| α9β1 |
Lymphangiogenesis |
| αIIbβ3 | Thrombus formation |
| α6β4 | Integrity of skin |
| αVβ3 | Suppresses tumorigenesis, angiogenesis, wound healing, inflammation and atherosclerosis |
| β2 integrins | Immune responses |