Actin Nucleation
Content
An Introduction to Actin Nucleation[Edit]
The polymerization of actin filaments requires numerous factors, however the fundamental unit is ATP-bound G-actin. The first step in actin polymerization is known as ‘nucleation’. This step sees the formation of an actin nucleus, which is essentially a complex of three actin monomers, from which an actin filament may elongate. Although non-muscle cells have a high concentration of G-actin-ATP (~100 μM) [1], pure G-actin monomers fail to nucleate new actin filaments efficiently due to the instability of actin oligomers. Additional factors are therefore required and although the exact mechanisms behind filament nucleation remain to be clearly defined, two models, each involving distinct mechanisms and proteins have been proposed. Importantly, these mechanisms are not mutually exclusive and it maybe be the case that nucleation of actin filaments results from a combination of both mechanisms [2].
 Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.
In the first model, known as the ‘tip nucleation model’, members of the formin family of proteins cluster at the plasma membrane and initiate the nucleation of actin filaments. Formin subsequently mediates filament extension with the structural integrity of the filament bundles maintained by fascin cross-linking [2]. The alternative model is known as the “convergent elongation model”. In this model, the Arp2/3 complex, which is more commonly associated with lamellipodia formation but has been found to be critical for filopodia initiation [3], plays a role. Here, Arp2/3 complex nucleated branches continually develop from the actin filament network located at the leading edge of the lamellipodia. These filaments are proposed to gradually converge, forming a bundle that is secured by facsin cross-linking.
Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.
In the first model, known as the ‘tip nucleation model’, members of the formin family of proteins cluster at the plasma membrane and initiate the nucleation of actin filaments. Formin subsequently mediates filament extension with the structural integrity of the filament bundles maintained by fascin cross-linking [2]. The alternative model is known as the “convergent elongation model”. In this model, the Arp2/3 complex, which is more commonly associated with lamellipodia formation but has been found to be critical for filopodia initiation [3], plays a role. Here, Arp2/3 complex nucleated branches continually develop from the actin filament network located at the leading edge of the lamellipodia. These filaments are proposed to gradually converge, forming a bundle that is secured by facsin cross-linking.
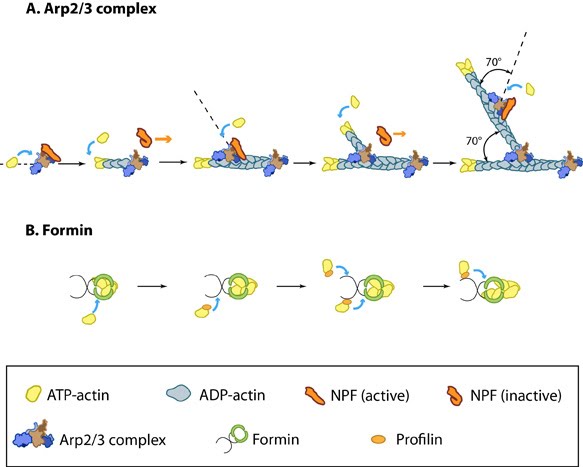 Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.
Figure 1. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.Formin mediates actin nucleation of unbranched filaments[Edit]
The formins are a large family of proteins that facilitate the nucleation of new, unbranched filaments by promoting the interaction between two actin monomers (reviewed in [4]). Under normal circumstances formins are auto-inhibited through structural interactions between the two ends of the protein [5]. However, conformational rearrangements resulting in their activation can be induced through interactions with GTP-bound (active) Rho GTPases [6]. This process remains poorly understood (reviewed in [7]).
 Figure 2. Formin-mediated nucleation of actin filaments: The FH2 domains of the formin dimer (shown in green)
bind to actin monomers to initiate filament assembly. Recent studies
indicate this is assisted, or even mediated, by additional factors such
as APC. The FH1 domains
of the formin dimer (shown as black lines) have short polyproline
sequences that interact with profilin. Profilin binds to both formin and
actin monomers to increase the addition of actin monomers to the barbed
end of the filament.In a recent study, the tumor suppressor adenomatous polyposis coli (APC) was shown to bind the formin mDia1 and overcome capping protein- and profilin-mediated suppression of spontaneous actin nucleation, resulting in the initiation of actin filament nucleation and elongation [8]. In the mechanism described, APC is primarily responsible for actin monomer recruitment, whilst mDia1 catalyzes filament elongation. Actin recruitment by APC did not involve capturing F-actin intermediates that had spontaneously formed nor did APC contribute to filament elongation. In this model, once actin polymerization commences the APC-mDia1 complex separates – mDia1 is propelled away from APC along with the growing barbed end of the filament and APC remains attached to the filament at the site of nucleation [8].
Figure 2. Formin-mediated nucleation of actin filaments: The FH2 domains of the formin dimer (shown in green)
bind to actin monomers to initiate filament assembly. Recent studies
indicate this is assisted, or even mediated, by additional factors such
as APC. The FH1 domains
of the formin dimer (shown as black lines) have short polyproline
sequences that interact with profilin. Profilin binds to both formin and
actin monomers to increase the addition of actin monomers to the barbed
end of the filament.In a recent study, the tumor suppressor adenomatous polyposis coli (APC) was shown to bind the formin mDia1 and overcome capping protein- and profilin-mediated suppression of spontaneous actin nucleation, resulting in the initiation of actin filament nucleation and elongation [8]. In the mechanism described, APC is primarily responsible for actin monomer recruitment, whilst mDia1 catalyzes filament elongation. Actin recruitment by APC did not involve capturing F-actin intermediates that had spontaneously formed nor did APC contribute to filament elongation. In this model, once actin polymerization commences the APC-mDia1 complex separates – mDia1 is propelled away from APC along with the growing barbed end of the filament and APC remains attached to the filament at the site of nucleation [8].
Although a consensus has yet to be reached for the mechanism of formin-mediated nucleation, it is now well-established that activated formins function as dimers and form a donut-shaped complex around terminal actin subunits, orientating themselves toward the (+) end of the actin filament or nucleus [9]. This binding is facilitated by FH2 (formin homology 2) domains within the formin monomers. Next, each formin monomer binds and captures profilin units, which are themselves already bound to G-actin monomers. This interaction is mediated by multiple stretches of polyproline residues within the FH1 domain of formins [10]. This domain is known to range from 15-229 residues, consist of between 35% and 100% proline residues, and contain up to 16 profilin binding sites [11]. Profilin maintains a steady pool of actin monomers by promoting ADP to ATP nucleotide exchange on G-actin[9]. These monomers of ATP-G-actin are then added the growing actin filament. The coupling of formin with the growing end prevents capping and allows continued growth of the filaments [12].
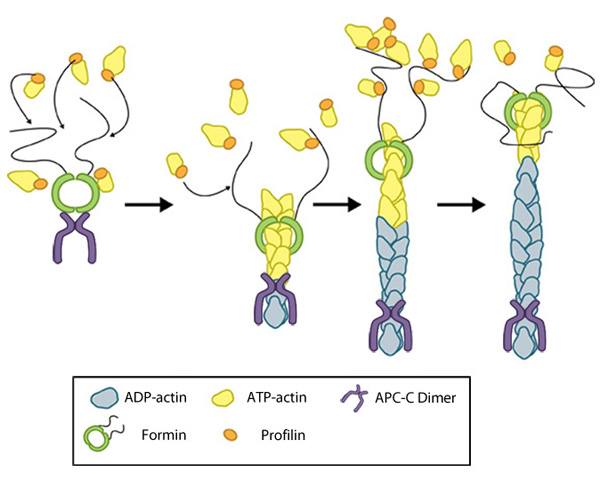 Figure 2. Formin-mediated nucleation of actin filaments: The FH2 domains of the formin dimer (shown in green)
bind to actin monomers to initiate filament assembly. Recent studies
indicate this is assisted, or even mediated, by additional factors such
as APC. The FH1 domains
of the formin dimer (shown as black lines) have short polyproline
sequences that interact with profilin. Profilin binds to both formin and
actin monomers to increase the addition of actin monomers to the barbed
end of the filament.
Figure 2. Formin-mediated nucleation of actin filaments: The FH2 domains of the formin dimer (shown in green)
bind to actin monomers to initiate filament assembly. Recent studies
indicate this is assisted, or even mediated, by additional factors such
as APC. The FH1 domains
of the formin dimer (shown as black lines) have short polyproline
sequences that interact with profilin. Profilin binds to both formin and
actin monomers to increase the addition of actin monomers to the barbed
end of the filament.Although a consensus has yet to be reached for the mechanism of formin-mediated nucleation, it is now well-established that activated formins function as dimers and form a donut-shaped complex around terminal actin subunits, orientating themselves toward the (+) end of the actin filament or nucleus [9]. This binding is facilitated by FH2 (formin homology 2) domains within the formin monomers. Next, each formin monomer binds and captures profilin units, which are themselves already bound to G-actin monomers. This interaction is mediated by multiple stretches of polyproline residues within the FH1 domain of formins [10]. This domain is known to range from 15-229 residues, consist of between 35% and 100% proline residues, and contain up to 16 profilin binding sites [11]. Profilin maintains a steady pool of actin monomers by promoting ADP to ATP nucleotide exchange on G-actin[9]. These monomers of ATP-G-actin are then added the growing actin filament. The coupling of formin with the growing end prevents capping and allows continued growth of the filaments [12].
Arp2/3-mediated nucleation of branched filaments[Edit]
The Arp2/3 complex is composed of 7 evolutionarily conserved subunits (Arp2, Arp3, ARPC1-C5) that are structurally similar to the barbed end of actin [13]. The complex is inherently inactive, however once activated it facilitates the nucleation of actin monomers from existing filaments as new branches or daughter filaments (reviewed in [14]).
Video: Arp2/3-mediated Actin Nucleation. The process by which the Arp2/3 complex mediates actin nucleation is shown here. This process is regulated by proteins such as WASp and Cdc42, which are depicted in this animation as stylized schematic models. [Video produced by MBInfo]
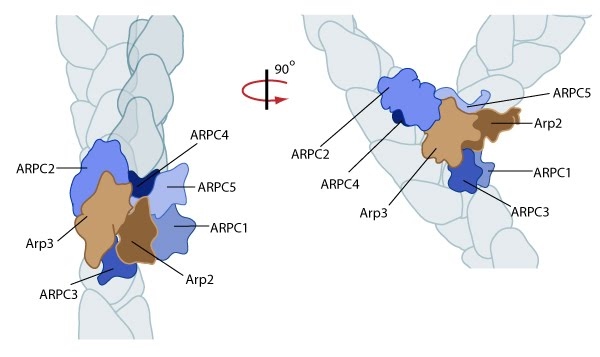 Figure 3. Reconstruction of Arp2/3 complex-mediated nucleation: Arp2/3 complex initiates a new branched filament by binding to the side of the mother filament and recruiting actin monomers. Components of the Arp2/3 complex remain at the pointed end of the filament (Figure adapted from [17]).
Figure 3. Reconstruction of Arp2/3 complex-mediated nucleation: Arp2/3 complex initiates a new branched filament by binding to the side of the mother filament and recruiting actin monomers. Components of the Arp2/3 complex remain at the pointed end of the filament (Figure adapted from [17]).The exact mechanism of nucleation remains unclear however it is believed that all seven subunits coordinate to anchor the pointed end of the new filament at a 78° angle to the existing actin network [17]. NPFs act to deliver actin subunits to the Arp2/3 complex at the barbed end, via binding through their WH2 domain – this serves to push the leading edge of a cell forward [22]. It has also been suggested that each component of the complex plays specific roles, with Arp2 and Arp3 interacting with the pointed end of the daughter filament and ARPC2 and ARPC4 binding to the original filament [17].
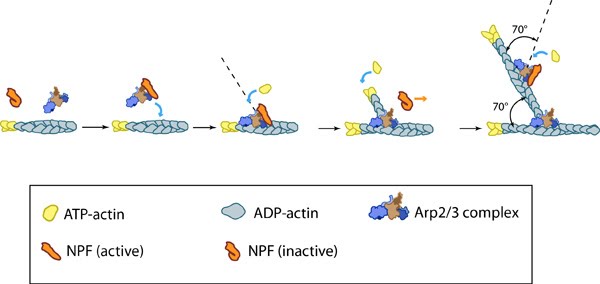 Figure 4. Arp2/3-mediated actin polymerization: NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate actin filaments that form new branches from the side of preexisting filaments. The Arp2/3 complex remains at the minus end of the filament.
Figure 4. Arp2/3-mediated actin polymerization: NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate actin filaments that form new branches from the side of preexisting filaments. The Arp2/3 complex remains at the minus end of the filament.This bias of Arp2/3 towards branch formation on convex curvature was proposed to represent a mechanosensing mechanism whereby detection of filament curvature alters the local cytosekelton in order to reinforce it against the imposing forces [23].
Tandem-monomer-binding nucleators[Edit]
Although the most commonly described nucleators are the Arp2/3 complex, and the formins, however a third group has also been identified. This third group is known as ‘tandem-monomer-binding nucleators’ as each member possesses tandem repeats of G-actin binding motifs.
Included in this group of nucleators are the Spire proteins, Cordon-bleu (Cobl), Leiomodin (Lmod-2), JMY and adenomatous polyposis coli (APC). Common to each of these proteins are repeats of the actin binding motif Wiskott-Aldrich Syndrome protein (WASp) homology 2 (WH2) domain. However, additional actin binding motifs may be present in the individual members, providing variation not their mechanisms of nucleation, but importantly, in the cellular functions they facilitate.
Animation used by permission. Source: Janet Iwasa