Formin-nucleated actin cable assembly[Edit]
Content
Formins promote the elongation of pre-existing filaments by removing barbed end capping proteins and forming a sleeve around the actin subunits. Formins are also capable of actin nucleation, a process which is spatiotemporally coupled with actin disassembly [1].
Formins nucleate and polymerize actin filaments at focal adhesions at a rate of around 0.3 µm/min [2]. Inhibiting formin protein expression results in a decreased filament elongation rate (0.1 µm/min), coupled with abnormal stress fiber morphology and an accumulation of actin binding proteins (e.g. α-actinin [2]). Ena/VASP proteins support formin-mediated filament elongation by tethering the filaments near sites of active actin assembly [3, 4].
Formins could, in theory, contribute to protrusive forces by remaining attached to the barbed end of actin filaments [5, 6] (reviewed in [7]). Consistent with this notion, excessive formin activity promotes cell migration. However precisely how this is achieved is unknown given that formin-induced activity does not impact the overall adherence of cells to their substrate, nor does it change the avidity or affinity of cell adhesion receptors (e.g. integrins) [8].
Formins nucleate and polymerize actin filaments at focal adhesions at a rate of around 0.3 µm/min [2]. Inhibiting formin protein expression results in a decreased filament elongation rate (0.1 µm/min), coupled with abnormal stress fiber morphology and an accumulation of actin binding proteins (e.g. α-actinin [2]). Ena/VASP proteins support formin-mediated filament elongation by tethering the filaments near sites of active actin assembly [3, 4].
Formins could, in theory, contribute to protrusive forces by remaining attached to the barbed end of actin filaments [5, 6] (reviewed in [7]). Consistent with this notion, excessive formin activity promotes cell migration. However precisely how this is achieved is unknown given that formin-induced activity does not impact the overall adherence of cells to their substrate, nor does it change the avidity or affinity of cell adhesion receptors (e.g. integrins) [8].
The Structure of Formin[Edit]
Actin filament assembly and disassembly are primary molecular processes that facilitate whole cell motility and the movement of subcellular structures. The initial stage in the formation of actin filaments is nucleation.
 Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domain
Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domain
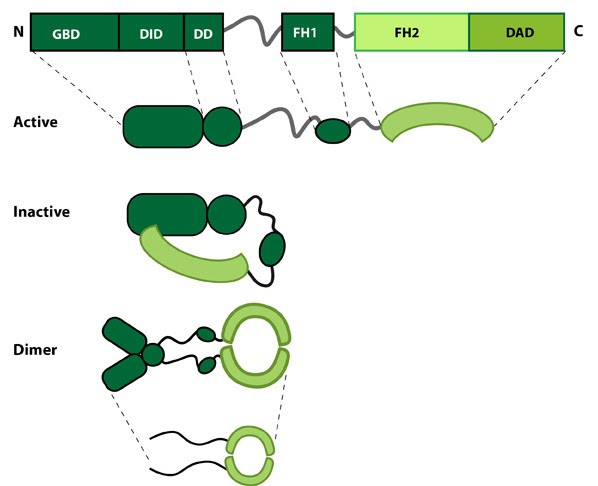 Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domain
Figure 1. Formin: Formin.This schematic diagram illustrates the molecular organization of formin. Relevant domains/regions that are believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [1]. An intramolecular interaction between the diaphanous inhibitory domain (DID)and the diaphanous auto-regulatory domain (DAD), which prevents formin (e.g. mDia1) from nucleating actin filaments, is relieved by Rho binding to the GTPase binding domain (GBD, aka CRIB domain) [2]. However, this regulation may be more complex [3, 4]. DD = dimerization domain, FH = formin homology domainNucleation is defined as the formation of a stable actin polymer from its monomeric units [5] and may also be facilitated by formin. Further to nucleation formins also facilitate the processive elongation of actin filaments, exclusively at the barbed end (reviewed in [6]). All members of this family share a common Formin homology 2 domain (FH2) and all but one (Dictyostelium discoideum formin, ForC) feature a FH1 domain. The diaphanous auto-regulatory domain (DAD), diaphanous inhibitory domain (DID) and the GTPase binding domain are also commonly identified in members of the formin family, though not found in all members. Each formin functions as a homodimer that shares structural similarity to F-actin. Each of these domains are functionally important, playing crucial roles in either the translocation of the protein along the growing actin filament, addition of actin monomers to the actin filament or in the regulation of the protein.
FH2
The FH2 domain is approximately 400 amino acids in length and binds to the barbed ends of actin filaments. Binding occurs following the dimerization of two arched FH2 domains, in a head-to-tail orientation. This produces a donut-shaped dimer held together by the interactions of the ‘lasso’ subdomain, within one of the FH2 domains, and the ‘post’ subdomain of the other FH2 domain. The linker region between these subdomains contains highly conserved residues that enable the nucleation and processive capping activity of formin. With a diameter of approximately 11nm, the donut shaped dimer easily accommodates an actin filament which is approximately 8nm wide. It has long been accepted that formins track along growing actin filaments without needing to dissociate and re-associate. Several models have been proposed to explain this movement (reviewed in [7]) with the most recent evidence suggesting the FH2 dimer will rotate according to the helical structure of F-actin [8].FH1
The FH1 domain is proline-rich and is associated with a high level of variation amongst the formin family. It varies in length from 15 to 229 residues and in proline content from 35% to 100%. It also contains a varying number of profilin binding sites [7], as exemplified in the human formins FHOD1 and FMNL2, with FHOD1 containing 2-3 profilin binding sites and FMNL2 containing 33. Although FH1 and FH2 are found in all formins (note the earlier exception above), it remains unclear whether co-operation between the domains is essential for formin function. It is generally accepted that a more concentrated pool of profilin–actin in close proximity to the FH2 domain increases the rate of elongation and that this may be further enhanced by the delivery of actin monomers by the FH1 domain to the barbed end of the filament in the correct orientation.DAD
Recently the DAD domain was implicated in the nucleation of actin monomers, despite not being present in all members of the formin family [9]. This domain (which lies at the C-terminal end of the molecule) was originally believed to function solely in the auto-inhibition of the protein, via its interaction with the N-terminal FH3 domain. The nucleation of actin monomers is believed to result from direct binding of the DAD domain with G-actin. This is suggested to be of particular importance in the nucleation of new filaments, the elongation of filaments in the absence of profilin and at very high concentrations of free profilin that would otherwise competitively block the FH1 domain [9].FH3 Domain (DID and DD)
The FH3 domain lies at the N-terminal end of the protein and is the least conserved domain among the various formins. It is important in the auto-regulation of the protein, forming a surface region that is recognized by the DAD domain. It is believed that binding of DAD to the FH3 domain renders the protein inactive and unable to dimerize or bind actin filaments. This however has yet to be confirmed experimentally.How do formins facilitate filament polymerization?[Edit]
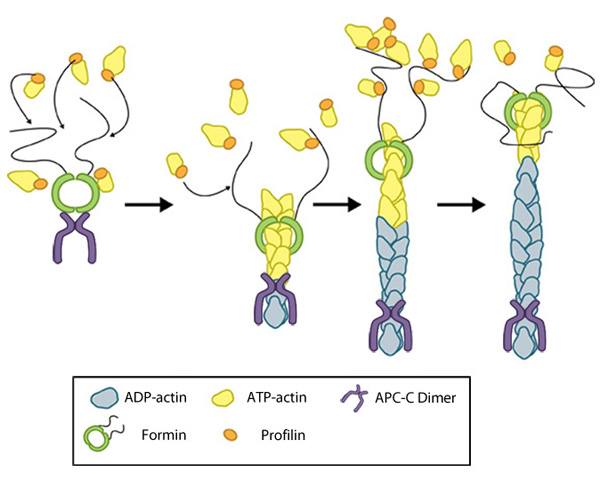 Figure 2. Formin-mediated nucleation of actin filaments: The FH2 domains of the formin dimer (shown in green)
bind to actin monomers to initiate filament assembly. Recent studies
indicate this is assisted, or even mediated, by additional factors such
as APC. The FH1 domains
of the formin dimer (shown as black lines) have short polyproline
sequences that interact with profilin. Profilin binds to both formin and
actin monomers to increase the addition of actin monomers to the barbed
end of the filament.
Figure 2. Formin-mediated nucleation of actin filaments: The FH2 domains of the formin dimer (shown in green)
bind to actin monomers to initiate filament assembly. Recent studies
indicate this is assisted, or even mediated, by additional factors such
as APC. The FH1 domains
of the formin dimer (shown as black lines) have short polyproline
sequences that interact with profilin. Profilin binds to both formin and
actin monomers to increase the addition of actin monomers to the barbed
end of the filament.Next, each formin monomer binds and captures profilin units, which are themselves already bound to G-actin monomers. This interaction is mediated by multiple stretches of polyproline residues within the FH1 domain of formins [13]. This domain is known to range from 15-229 residues, consist of between 35% and 100% proline residues, and contain up to 16 profilin binding sites [7]. Profilin maintains a steady pool of actin monomers by promoting ADP to ATP nucleotide exchange on G-actin[14]. These monomers of ATP-G-actin are then added the growing actin filament. The coupling of formin with the growing end prevents capping and allows continued growth of the filaments [15].
The role of profilin in formin-nucleated actin cable assembly[Edit]
Profilin binds simultaneously to formin and actin monomers; this interaction tethers multiple profilin-actin complexes near the growing end of actin filaments, which promotes the processive addition of actin subunits [16, 17]. Profilin uses the energy from ATP hydrolysis generated during actin polymerization to facilitate actin assembly [17]. Profilin binds to cytoplasmic ATP-actin monomers better than cytoplasmic ADP-actin monomers [18].
Profilin has been suggested to generally increase the elongation rate of formin-associated filaments by:
* Catalyzing the exchange of ADP for ATP on actin monomers [19, 20].
* Blocking free monomers from elongating pointed ends [19].
* Lowering the critical concentration at the barbed end [17].
* Promoting the association of G-actin-ATP to the barbed end [21].
* Catalyzing the exchange of ADP for ATP on actin monomers [19, 20].
* Blocking free monomers from elongating pointed ends [19].
* Lowering the critical concentration at the barbed end [17].
* Promoting the association of G-actin-ATP to the barbed end [21].
Importantly however, profilin also promotes disassembly of actin filaments by sequestering monomeric G-actin, thereby blocking its association with the barbed ends and promoting its disassembly from the pointed ends of actin filaments [19]. The combined actions of profilin and ADF/cofilin synergize to enhance turnover of actin filaments [22].