Actin filament polymerization
Content
- Actin Filaments (F-actin) grow from the polymerization of G-actin monomers
- How do actin monomers polymerize to form an actin filament?
- Dynamics and Components for Actin Filament Polymerization
- Nucleation Promoting Factors and NPF Accessory Proteins regulate actin filament polymerization
- Ena/VASP
- Cortactin
Actin Filaments (F-actin) grow from the polymerization of G-actin monomers[Edit]
Actin is a highly abundant (10-100 micromolar on average),~42 kDa structural protein found in all eukaryotic cells (except for nematode sperm). With more than 95% conservation in the primary structure, actin is one of the most highly-conserved proteins [1]. The monomeric, globular form of actin, known as G-actin, forms the basic unit for actin filaments. In many cases actin filaments may bundle together with other actin filaments, or, together with their associated motor proteins (e.g. myosin superfamily) form an elaborate network known as the actin cytoskeleton. This occurs primarily at or near the plasma membrane. Consequently a region of high actin filament density is commonly found at the cell periphery and is known as the cell cortex. Actin filaments in the cell cortex determine the shape, stiffness and movement of the cell surface. The actin cytoskeleton also facilitates the transduction of mechanical signals, and can generate the intracellular forces that are required for many cellular functions including cell motility, muscle contraction, cell division, cytokinesis, vesicle and organelle movement and cell signaling. Actin also contributes to the formation and maintenance of cell junctions.
 Figure 1. Structure of G-actin and its assembly into filaments: The structure shown here [2] was downloaded from the RSCB Protein Data Bank (PDB file: 1atn). The ATP binding cleft is starred (*) on the right. Actin comprises four subdomains, termed SD1 (blue), SD2 (red), SD3 (grey) and SD4 (green). The barbed end of each monomer (SD1 and SD3) is shown on the left and the pointed end (SD2 and SD4) is shown on the right. Similarly the polarity of the actin filament, which comprises these monomers is shown at the bottom, with the barbed (+) end on the left and the pointed (-) end on the right.Higher eukaryotes commonly express several isoforms of actin; encoded by a family of related genes. Actin isoforms are divided into three classes (alpha [α], beta [β] and gamma [γ]) according to their isoelectric point. In general, alpha actins are found in muscle (α-skeletal, α-aortic smooth, α-cardiac, and γ2-enteric smooth), whereas beta and gamma isoforms are prominent in non-muscle cells (β- and γ1-cytoplasmic). Early models that described actin filaments were constructed by fitting the filament x-ray crystal structure to the atomic structure of actin monomers [3] (reviewed in [4]). More recent models used a number of different approaches [5,6]. Collectively however these results suggest that when single actin strands form, two asymmetric actin monomers align to form a twofold axis of symmetry [7]; their subsequent assembly into a filament that is composed of a pair of strands causes a left-handed helical twist when the adjacent subunits are positioned with respect to each other [8].
Figure 1. Structure of G-actin and its assembly into filaments: The structure shown here [2] was downloaded from the RSCB Protein Data Bank (PDB file: 1atn). The ATP binding cleft is starred (*) on the right. Actin comprises four subdomains, termed SD1 (blue), SD2 (red), SD3 (grey) and SD4 (green). The barbed end of each monomer (SD1 and SD3) is shown on the left and the pointed end (SD2 and SD4) is shown on the right. Similarly the polarity of the actin filament, which comprises these monomers is shown at the bottom, with the barbed (+) end on the left and the pointed (-) end on the right.Higher eukaryotes commonly express several isoforms of actin; encoded by a family of related genes. Actin isoforms are divided into three classes (alpha [α], beta [β] and gamma [γ]) according to their isoelectric point. In general, alpha actins are found in muscle (α-skeletal, α-aortic smooth, α-cardiac, and γ2-enteric smooth), whereas beta and gamma isoforms are prominent in non-muscle cells (β- and γ1-cytoplasmic). Early models that described actin filaments were constructed by fitting the filament x-ray crystal structure to the atomic structure of actin monomers [3] (reviewed in [4]). More recent models used a number of different approaches [5,6]. Collectively however these results suggest that when single actin strands form, two asymmetric actin monomers align to form a twofold axis of symmetry [7]; their subsequent assembly into a filament that is composed of a pair of strands causes a left-handed helical twist when the adjacent subunits are positioned with respect to each other [8].
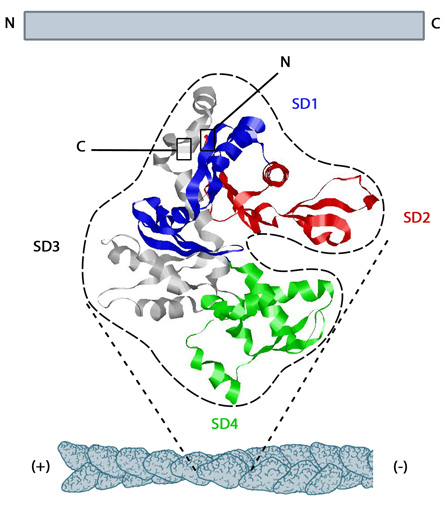 Figure 1. Structure of G-actin and its assembly into filaments: The structure shown here [2] was downloaded from the RSCB Protein Data Bank (PDB file: 1atn). The ATP binding cleft is starred (*) on the right. Actin comprises four subdomains, termed SD1 (blue), SD2 (red), SD3 (grey) and SD4 (green). The barbed end of each monomer (SD1 and SD3) is shown on the left and the pointed end (SD2 and SD4) is shown on the right. Similarly the polarity of the actin filament, which comprises these monomers is shown at the bottom, with the barbed (+) end on the left and the pointed (-) end on the right.
Figure 1. Structure of G-actin and its assembly into filaments: The structure shown here [2] was downloaded from the RSCB Protein Data Bank (PDB file: 1atn). The ATP binding cleft is starred (*) on the right. Actin comprises four subdomains, termed SD1 (blue), SD2 (red), SD3 (grey) and SD4 (green). The barbed end of each monomer (SD1 and SD3) is shown on the left and the pointed end (SD2 and SD4) is shown on the right. Similarly the polarity of the actin filament, which comprises these monomers is shown at the bottom, with the barbed (+) end on the left and the pointed (-) end on the right.How do actin monomers polymerize to form an actin filament?[Edit]
Actin filaments are highly dynamic and their polymerization is usually correlated to their disassembly. Generally actin filament polymerization occurs over three phases: A nucleation phase, an elongation phase and a steady state phase.
Step 3: Steady State Phase (Treadmilling)
 Figure 3. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the Cc
Figure 3. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the Cc
When the association rate of free ATP-G-actin is greater than the rate of subunit loss, the filament appears to grow, creating a ‘cap’ rich in ATP-subunits [10]. Conversely, when the association rate of free ATP-actin is lower than the rate of subunit loss, the filament is seen to shrink. When the association rate of free ATP-actin is equal to the rate of dissociation at the (-) end, no net growth occurs and this is known as ‘treadmilling’.
Table 2: Cellular concentrations (in µM) of key proteins in the actin system of diverse cells (if known, modified from [10] and [11]). * These values are representative of biological concentrations similar to those used for in vitro reconstitution experiments of bacterial motility and so are not cell type specific[11].
During the nucleation phase the formation of a stable ‘actin nucleus’ occurs. This is usually comprised of three actin monomers in complex. In the elongation phase monomers are rapidly added to the filament at the (+ve) or barbed end and this is often facilitated by additional elongation factors such as formin. In the steady state phase, the filament dynamics enter a state of equilibrium where monomer disassembly from the (-) end and polymerization at the (+) end is balanced and maintained by a critical concentration of monomers in the cytosol. This steady state assembly and disassembly is known as ‘treadmilling’.
Video: Actin filament assembly. The actin network is made up of filamentous actin (F-actin). These filaments are highly dynamic in nature and comprise monomers of G-actin bound to either ATP (yellow) or ADP (blue). Assembly is powered by ATP hydrolysis and filament nucleation happens spontaneously in vitro. Polymerization: Addition of ATP-actin occurs at the barbed end, leading to filament elongation. Elongation will continue whilst the rate of elongation is greater than the loss of ADP-actin from the pointed end. Profilin preferentially binds to ATP-actin, inhibits nucleation and accelerates filament elongation in vivo. Depolymerization: When the dissociation rate of ADP-actin exceeds the rate of ATP-actin association, the filament shrinks. In vivo, this is aided by cofilin, which can severe filaments into short fragments and promote subunit loss from the pointed ends. Actin treadmilling occurs when the rate of association of ATP-actin and the rate of loss of ADP-actin are balanced. [Video produced by MBInfo]
 Figure 2. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.Step 1: Actin Nucleation
Figure 2. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.Step 1: Actin Nucleation
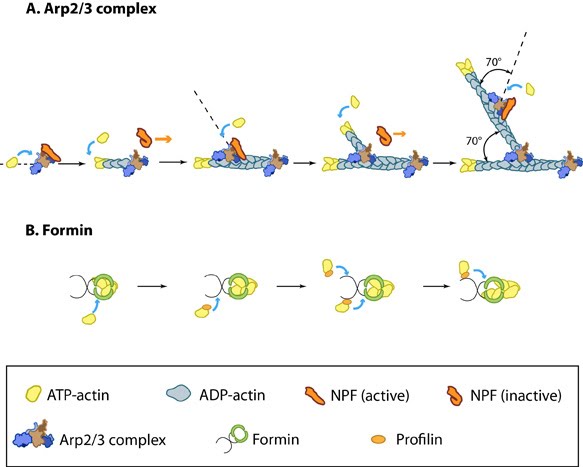 Figure 2. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.
Figure 2. Accessory proteins nucleate actin filaments.: A. NPFs (e.g. WASp; Scar) bring together the Arp2/3 complex and actin monomers to nucleate new actin filaments and to form new branches from the side of pre-existing filaments. Arp2/3 complex remains at the minus end of the filament. B. Formin cooperates with profilin to nucleate new actin filaments. Formin remains at the plus end of the filament.The first step in actin polymerization is known as ‘nucleation’. This step sees the formation of an actin nucleus, which is essentially a complex of three actin monomers, from which an actin filament may elongate. Although actin monomers will spontaneously oligomerize in solution when present at a concentration above the critical concentration (Cc), these complexes are highly unstable. Actin nucleation therefore requires additional proteins known as ‘actin nucleators’ to promote the formation of a stable actin nucleus. These proteins include the Arp2/3 complex, formins, and the ‘tandem-monomer-binding nucleators’.
Step 2: Filament Elongation Phase
In the 2nd step, the rapid addition of actin monomers to the (+ve) end of the actin filament occurs. This process, known as elongation is mediated by proteins that translocate along the growing filament and simultaneously catalyze the addition of monomers to the filament end. Such proteins include members of the formin family of proteins. For this process to occur, the (+) end of the filament must be exposed, and this means removal of capping protein.
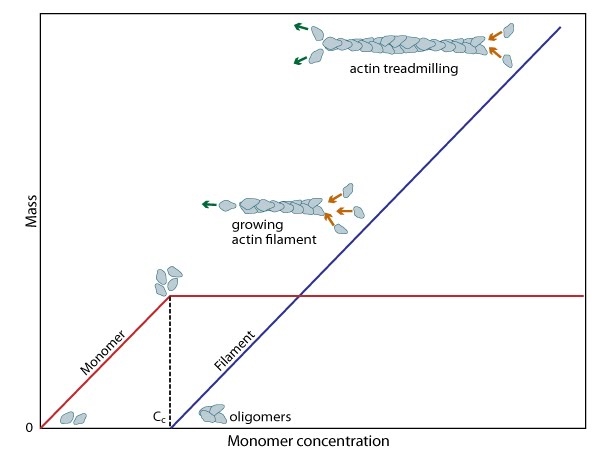 Figure 3. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the Cc
Figure 3. Cc and actin filament assembly.: The critical concentration (Cc) marks the level at which G-actin monomers are in equilibrium with the actin filaments. Actin filaments are only formed at monomer concentrations above the CcA steady pool of actin monomers must be maintained to enable a polymerization to continue beyond the rapid elongation phase. This is, in part, aided by the actin binding protein profilin, which promotes ADP to ATP nucleotide exchange on G-actin. However, the rate of monomer dissociation from the (-) end of the filament is also important. Dissociation of the subunits ultimately results from ATP hydrolysis, which induces a conformational change in the actin subunit that weakens its association with neighboring subunits (as reviewed in [8]). The concentration of actin monomers in the cytosol will either favor disassembly, or assembly of the actin filament,
and these values are known as the critical concentration (Cc). When the concentration of free subunits exceeds the Cc, filament elongation occurs spontaneously [9]. Importantly, the Cc usually varies between the filament (+) end and the (-) end. At the steady state, which is achieved when the rate of filament polymerization is equally balanced by filament disassembly, the free subunit concentration is higher than the Cc at the (+) end and lower than the Cc at the (-) end. This results in subunits being added to the (+) end and dissociating from the (-).
When the association rate of free ATP-G-actin is greater than the rate of subunit loss, the filament appears to grow, creating a ‘cap’ rich in ATP-subunits [10]. Conversely, when the association rate of free ATP-actin is lower than the rate of subunit loss, the filament is seen to shrink. When the association rate of free ATP-actin is equal to the rate of dissociation at the (-) end, no net growth occurs and this is known as ‘treadmilling’.
| Cc | Rate of G-actin addition | Rate of G-actin dissociation | |
| (-) end | 0.6 µM | 1.3 µM -1 s -1 | 0.8 s -1 |
| (+) end | 0.12 µM | 12 µM -1 s -1 | 1.4 s -1 |
| Protein | Acanthamoeba | Dictyostelium | Xenopusegg extract | S. cerevisiae | in silico Listeriamodel * |
| Polymerized actin (e.g. F-actin) | 100 | 90 | 4 | 2 | |
| Unpolymerized actin | 00 | 160 | 12 | 0.01 | 12 |
| Profilin | 100 | 5 | present | 5 |
|
| Thymosin-β4 | absent? | absent? |
20 | absent | |
| ADF/cofilin | 20 | <100 | 3 | present | 3 |
| Arp2/3 complex | 2-4 | present | present | 0.3 |
|
| Capping protein | 1 | 1 | 1 | 1 |
|
| Gelsolin | ? absent | ||||
| α-actinin | 4 | 3 | |||
| VASP | 0.5 |
||||
| Act A |
105/µm2 |
Dynamics and Components for Actin Filament Polymerization[Edit]
Actin filaments are dynamic structures whose growth and disassembly are tightly controlled by additional proteins. These proteins may either promote actin filament nucleation by stabilizing the actin nucleus, catalyze filament elongation or promote actin treadmilling. Some well established proteins that play such roles include the Arp2/3 complex, which facilitates nucleation of filament branches; profilin, which catalyzes ADP to ATP exchange and ADF/cofilin, which mediates filament disassembly. The cooperation between each component is extensive and each element has an optimal concentration.
 Figure 4. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
Figure 4. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
Factors influencing actin filament length and treadmilling
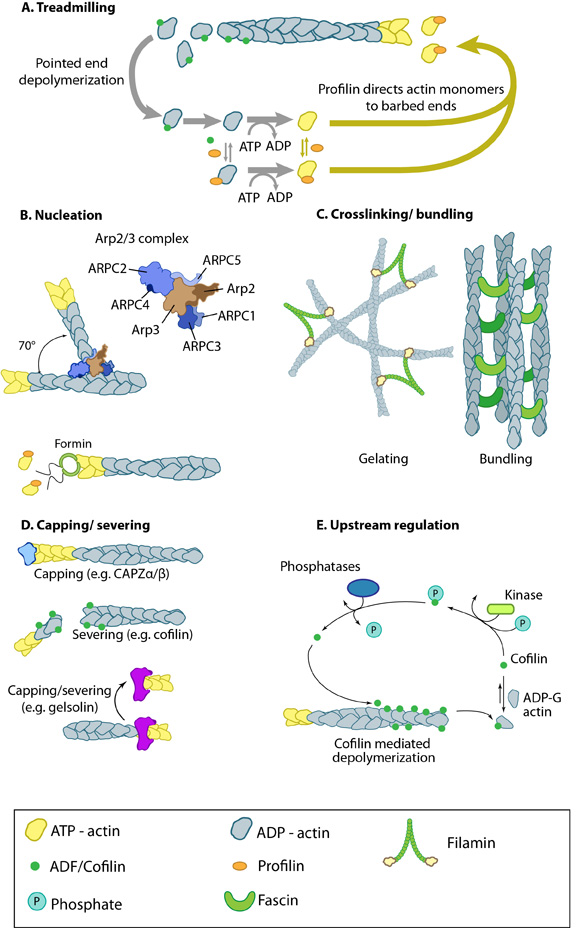 Figure 4. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.
Figure 4. Actin binding proteins influence actin dynamics: A. Treadmilling of actin filaments can be altered by profilin and ADF which generally increase and decrease the size of actin filaments, respectively. B. New filaments are nucleated by the ARP2/3 complex, which binds both G-actin monomers and the side of actin filaments to nucleate new filaments or branches. Formins nucleate new filaments by binding G-actin and through cooperation with profilin. C. Actin cross-linking proteins influence the packing and organization of actin filaments into secondary structures. D. Capping and severing proteins promote disassembly of actin filaments. E. Actin filament assembly can be modulated by events such as controlled nucleotide hydrolysis (e.g. ATP on actin) and reversible modifications (e.g. phosphorylation) on components that control actin assembly.1. ATP binding on G-actin and free ATP-G-actin concentration
ATP-binding on actin subunits modulates the dynamics of filament assembly, with ATP-binding generally favoring intersubunit interactions and thereby filament assembly [9]. The rate of actin addition to filaments depends on the concentration of free ATP-G-actin whereas the rate of subunit loss does not. At high free ATP-G-actin concentrations the rate of addition exceeds the rate of dissociation and this results in actin filament growth.2. The rate of ATP-G-actin assembly to the ends
The addition of ATP-G-actin to the two ends of preexisting actin filaments occurs at very different rates (see Table 1) [12]. The addition of free ATP-G-actin at the (-) end is much lower relative to the (+) end.3. The critical concentration can be adjusted
The (-) and (+) ends have a different criticial concentration (Cc) for actin filament growth. The Cc is defined as the concentration level of free ATP-G-actin where the rate of addition is balanced by the rate of loss and no net growth occurs at that end. At concentrations above the Cc, actin filament growth occurs, wheras below it, there is a loss of subunits and shrinkage occurs. Any protein that alters the Cc, e.g. profilin, will alter actin filament dynamics.4. Actin binding drugs
Toxins such as phalloidins, cytochalasins, latrunculin A, and jasplakinolide are naturally occurring small molecules that bind to actin and alter its polymerization. Phalloidins inhibit actin filament disassembly by locking adjacent actin subunits together, while cytochalasins bind to the barbed end of actin filaments to prevent actin filament assembly and disassembly at that end. Latrunculin A binds to actin monomers to inhibit their polymerization and thus promotes filament disassembly; latrunculin A may also inhibit nucleotide exchange on actin subunits [13, 14]. Jasplakinolides stabilize actin monomers, thereby enhancing filament nucleation and assembly.5. Actin binding proteins
Certain actin binding proteins initiate actin filament assembly/disassembly directly, while others influence the ATP binding, the rate of G-actin assembly and the Cc of the filament ends. There are over 60 families of actin-binding proteins (reviewed in [6]) with the main actin monomer-binding proteins in vertebrate cells being thymosin-β4 and profilin. Thymosin-β4 binds strongly to ATP-actin and prevents its assembly into filaments [15]. Because profilin and thymosin-β4 have overlapping binding sites on actin [16, 17], profilin must compete with thymosin-β4 during filament assembly [18]. Other examples of actin-binding proteins include: the nucleators spire and the Arp2/3 complex, elongation factors such as formin, actin cross-linking proteins such as fascin or filamin A, nucleation promoting factors such as WASp, and capping protein.Nucleation Promoting Factors and NPF Accessory Proteins regulate actin filament polymerization[Edit]
Additional proteins are required for the initiation and regulation of actin filament polymerization. These include the Nucleation Promoting Factors (NPF) and NPF Accessory Proteins.
 Figure 5. WASP_WAVE_nucleation_promoting_factors: The molecular organization of Scar/WAVE family members and WASP, with examples of how the NPFs are represented in figures throughout this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [23, 24]). WHD= WAVE (also known as SCAR)-homology domain; WIP= WASP-interacting protein; V= verprolin-homology domain (i.e. WASP-homology-2 domain [WH2]); C= cofilin-homology domain; A= acidic-rich domain; WH1= WASP-homology-1 (aka Ena-VASP-homology-1 [EVH1]); GBD= GTPase binding domain. Of note: N-WASP contains an additional V domain, and the Scar/WAVE proteins lack a direct binding domain for the Rho GTPase family.Class I proteins are classified into five groups:
Figure 5. WASP_WAVE_nucleation_promoting_factors: The molecular organization of Scar/WAVE family members and WASP, with examples of how the NPFs are represented in figures throughout this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [23, 24]). WHD= WAVE (also known as SCAR)-homology domain; WIP= WASP-interacting protein; V= verprolin-homology domain (i.e. WASP-homology-2 domain [WH2]); C= cofilin-homology domain; A= acidic-rich domain; WH1= WASP-homology-1 (aka Ena-VASP-homology-1 [EVH1]); GBD= GTPase binding domain. Of note: N-WASP contains an additional V domain, and the Scar/WAVE proteins lack a direct binding domain for the Rho GTPase family.Class I proteins are classified into five groups:
1) Wiskott-Aldrich Syndrome protein (WASP) and neuronal-enriched homologue of WASP (N-WASP)
2) WASP family Verprolin-homologous (WAVE) proteins (aka Suppressor of cAMP receptor [Scar])
3) WASP and Scar homologue (WASH)
4) WASP homologue associated with actin, membranes and microtubules (WHAMM)
5) junction-mediating regulatory protein (JMY)
Class II includes proteins such as cortactin.
The conserved verpolin-cofilin-homology and acidic-rich (VCA) domain at the carboxy terminus of WASP and Scar family members binds directly to the Arp2/3 complex to increase its nucleation activity [25, 26, 27, 28]. WASp also associates with other signaling components (e.g. haemopoietic cell kinase) and formins to modulate actin polymerization for cell polarization and chemotaxis in neutrophils [29, 30]. WASp and formins also cooperate to control the balance between lamellipodial protrusion activity in epithelial cells [31]. The WASP family of NPFs are normally auto-inhibited due to protein interactions which prevent the NPF from associating with actin and the Arp2/3 complex; they are activated by Rho GTPases and PIP2 [26] (reviewed in [32, 33]). In contrast, the Scar/WAVE proteins have constitutive activity [34]. NPF accessory proteins also modulate the activity of NPFs.
Examples of NPF accessory proteins include Verprolin (yeast), which modulates the activity of WASp with type I myosins, to promote actin assembly by Arp2/3 complex [35]. WASp-interacting proteins (WIPs) will also regulate the WASp activity. For example WIP [36] not only inhibits N-WASP, but also promotes nucleation and activation of the Arp2/3 complex through the coordinated binding of actin and another NPF, cortactin [37]. SPIN90/WISH (SH3 protein interacting with Nck, 90 kDa/WASP-interacting SH3 protein) [38]) on the other hand increases actin assembly in dendritic filopodia/spines independently of N-WASP through its association with the neuron-specific scaffolding protein, PSD-95 [38]. Surprisingly, indirect evidence shows that WIPs are required for WASp function [39].
Nucleation Promoting Factors
NPFs (e.g. WASP, Scar/WAVE) modulate actin filament nucleation by bringing together actin monomers and pre-existing actin filaments, for example, during filopodial initiation where they recruit the Arp2/3 complex. NPFs compete with profilin for binding to free actin (which inhibits actin nucleotide exchange) [19,20, 21, 22]; these combined functions promote actin-filament assembly at the barbed end.
Mammalian NPFs are broadly grouped into 2 classes:
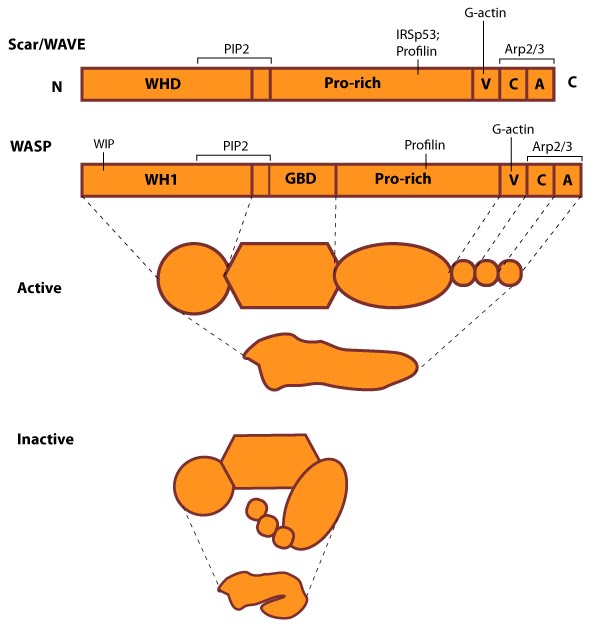 Figure 5. WASP_WAVE_nucleation_promoting_factors: The molecular organization of Scar/WAVE family members and WASP, with examples of how the NPFs are represented in figures throughout this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [23, 24]). WHD= WAVE (also known as SCAR)-homology domain; WIP= WASP-interacting protein; V= verprolin-homology domain (i.e. WASP-homology-2 domain [WH2]); C= cofilin-homology domain; A= acidic-rich domain; WH1= WASP-homology-1 (aka Ena-VASP-homology-1 [EVH1]); GBD= GTPase binding domain. Of note: N-WASP contains an additional V domain, and the Scar/WAVE proteins lack a direct binding domain for the Rho GTPase family.
Figure 5. WASP_WAVE_nucleation_promoting_factors: The molecular organization of Scar/WAVE family members and WASP, with examples of how the NPFs are represented in figures throughout this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [23, 24]). WHD= WAVE (also known as SCAR)-homology domain; WIP= WASP-interacting protein; V= verprolin-homology domain (i.e. WASP-homology-2 domain [WH2]); C= cofilin-homology domain; A= acidic-rich domain; WH1= WASP-homology-1 (aka Ena-VASP-homology-1 [EVH1]); GBD= GTPase binding domain. Of note: N-WASP contains an additional V domain, and the Scar/WAVE proteins lack a direct binding domain for the Rho GTPase family.1) Wiskott-Aldrich Syndrome protein (WASP) and neuronal-enriched homologue of WASP (N-WASP)
2) WASP family Verprolin-homologous (WAVE) proteins (aka Suppressor of cAMP receptor [Scar])
3) WASP and Scar homologue (WASH)
4) WASP homologue associated with actin, membranes and microtubules (WHAMM)
5) junction-mediating regulatory protein (JMY)
Class II includes proteins such as cortactin.
The conserved verpolin-cofilin-homology and acidic-rich (VCA) domain at the carboxy terminus of WASP and Scar family members binds directly to the Arp2/3 complex to increase its nucleation activity [25, 26, 27, 28]. WASp also associates with other signaling components (e.g. haemopoietic cell kinase) and formins to modulate actin polymerization for cell polarization and chemotaxis in neutrophils [29, 30]. WASp and formins also cooperate to control the balance between lamellipodial protrusion activity in epithelial cells [31]. The WASP family of NPFs are normally auto-inhibited due to protein interactions which prevent the NPF from associating with actin and the Arp2/3 complex; they are activated by Rho GTPases and PIP2 [26] (reviewed in [32, 33]). In contrast, the Scar/WAVE proteins have constitutive activity [34]. NPF accessory proteins also modulate the activity of NPFs.
NPF Accessory Proteins
Accessory proteins bind to WASp and Scar/WAVE proteins to modulate their NPF activity.Examples of NPF accessory proteins include Verprolin (yeast), which modulates the activity of WASp with type I myosins, to promote actin assembly by Arp2/3 complex [35]. WASp-interacting proteins (WIPs) will also regulate the WASp activity. For example WIP [36] not only inhibits N-WASP, but also promotes nucleation and activation of the Arp2/3 complex through the coordinated binding of actin and another NPF, cortactin [37]. SPIN90/WISH (SH3 protein interacting with Nck, 90 kDa/WASP-interacting SH3 protein) [38]) on the other hand increases actin assembly in dendritic filopodia/spines independently of N-WASP through its association with the neuron-specific scaffolding protein, PSD-95 [38]. Surprisingly, indirect evidence shows that WIPs are required for WASp function [39].
In another example, the WAVE complex will inhibit Scar/WAVE proteins from activating the Arp2/3 complex. The NPF activity of Scar/WAVE is restored during nucleation when Rac-GTP causes the dissociation of the WAVE complex from WAVE1 [40]. Similarly, IRSp53 has been implicated in both lamellipodia and filopodia formation/protrusion by augmenting the Rac-GTP-induced activation of WAVE NPF activity [41, 42].
Ena/VASP[Edit]
Proteins of the Ena/VASP family contribute to cell movement, axon guidance, neural tube closure and shape change in vertebrate cells by modulating actin filament organization and dynamics; these effects are achieved in part by regulating the morphology and behavior of actin-based structures such as lamellipodia and filopodia (reviewed in [43]). Ena/VASP proteins also modulate actin dynamics at sites of cell-ECM and cell-cell interactions and they are concentrated to the proximal portion of phosphotyrosine-rich domains at the ends of F-actin stress fibers [44]
Ena/VASP proteins promote actin filament elongation by tethering actin filaments to sites of active actin assembly [45, 46, 47]. Ena/VASP proteins recruit actin nucleation and initiation factors (e.g. Arp2/3 complex, formins) and promote F-actin assembly through profilin-binding (reviewed in [48]). The rate of Ena/VASP assisted actin filament elongation is determined by the recruitment of G-actin. This will occur via a G-actin binding site (GAB) that lies within the EVH2 domain and shares close sequence homology to WASP homology 2 motifs [49]. Ena/VASP proteins are also thought to accumulate at the plasma membrane where they alter actin polymerization by antagonizing the barbed (+) end capping proteins, thereby enabling the incorporation of actin into longer filaments [45, 46]; however, controversy over their exact mechanism still exists (reviewed in [43]). In addition, Ena/VASP may promote actin assembly by an unknown mechanism that is independent of initiation factors, however, this has not been demonstrated in intact cells [50]
Ena/VASP proteins promote actin filament elongation by tethering actin filaments to sites of active actin assembly [45, 46, 47]. Ena/VASP proteins recruit actin nucleation and initiation factors (e.g. Arp2/3 complex, formins) and promote F-actin assembly through profilin-binding (reviewed in [48]). The rate of Ena/VASP assisted actin filament elongation is determined by the recruitment of G-actin. This will occur via a G-actin binding site (GAB) that lies within the EVH2 domain and shares close sequence homology to WASP homology 2 motifs [49]. Ena/VASP proteins are also thought to accumulate at the plasma membrane where they alter actin polymerization by antagonizing the barbed (+) end capping proteins, thereby enabling the incorporation of actin into longer filaments [45, 46]; however, controversy over their exact mechanism still exists (reviewed in [43]). In addition, Ena/VASP may promote actin assembly by an unknown mechanism that is independent of initiation factors, however, this has not been demonstrated in intact cells [50]
Cortactin[Edit]
Cortactin is a class II nucleation promoting factor (NPF) that binds to actin filaments and influences their stability. Cortactin specifically stabilizes Arp2/3-mediated branch points along actin filaments through its repetitive actin binding sites [51] (reviewed in [52]). Although cortactin is a weak activator of the Arp2/3 complex when compared to class I NPFs (e.g. WASP, SCAR/WAVE), cortactin also binds to other NPFs (e.g. N-WASP) and their interacting proteins (e.g. WIP). This association may help to both recruit and activate Arp2/3 complex-mediated nucleation of actin filaments [37, 53, 54, 55, 56].
 Figure 6. Cortactin: This schematic diagram illustrates the molecular organization of cortactin (reviewed in [23])
and provides examples for how cortactin is represented in figures
throughout this resource. Relevant domains believed to be important for
cortactin activity are highlighted.Cortactin comprises five major domains [57]. The N-terminal acidic domain weakly binds and activates Arp2/3, via a DDW motif. The actin binding domain contains four to six actin binding repeats [58], with repeat four coordinating F-actin binding [58]. Each repeat is made up of a stretch of 37 amino acids. An SH3 domain (Src homology domain 3) facilitates binding of proteins containing a conserved proline-rich motif, such as Shank [59], dynamin-2 [60] and N-WASP [61].
Figure 6. Cortactin: This schematic diagram illustrates the molecular organization of cortactin (reviewed in [23])
and provides examples for how cortactin is represented in figures
throughout this resource. Relevant domains believed to be important for
cortactin activity are highlighted.Cortactin comprises five major domains [57]. The N-terminal acidic domain weakly binds and activates Arp2/3, via a DDW motif. The actin binding domain contains four to six actin binding repeats [58], with repeat four coordinating F-actin binding [58]. Each repeat is made up of a stretch of 37 amino acids. An SH3 domain (Src homology domain 3) facilitates binding of proteins containing a conserved proline-rich motif, such as Shank [59], dynamin-2 [60] and N-WASP [61].
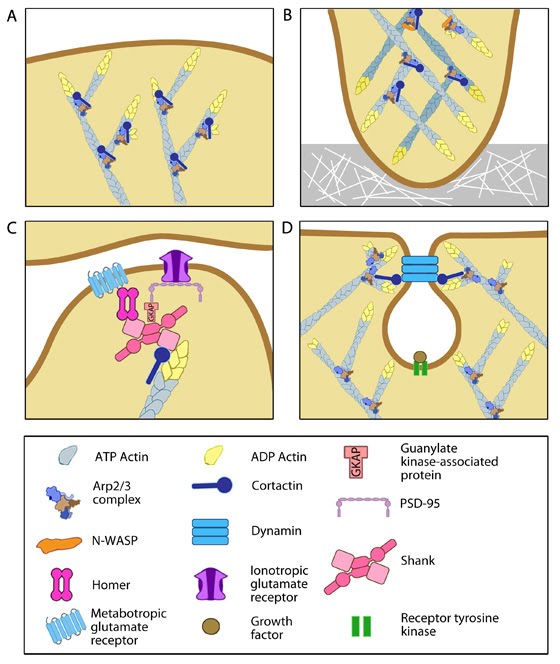
Functionally, cortactin is involved in a wide range of cellular processes pertaining to a variety of structures. The protein is highly enriched in mature neurons and is necessary for the formation and stability of dendritic spines [62]. It also associates with post synaptic densities (PSDs) of neurons expressing the ionotropic glutamate receptor, NMDA (N-methyl-D-aspartate). This association is via a weak interaction with the PSD protein Shank [59]. The function of cortactin at PSDs is not yet clear but it is speculated to facilitate changes in the actin cytoskeleton in response to synaptic activity. This is supported by the translocation of cortactin to synapses in response to stimulation by glutamate [59].
Cortactin also forms part of the actin-rich core of both podosomes and invadopodia [63]. Actin polymerization within invadopodia is essential to the function of this structure and the migratory behavior of cancerous cells [62]. Cortactin also promotes cell motility in normal cells and as such localizes to lamellipodia [64, 65]. It enhances lamellipodial persistence i.e. increases the time spent protruding, through its interactions with both Arp2/3 and F-actin. Cortactin demonstrates a notable preference for newly polymerized actin filaments i.e. those containing a greater amount of ATP-actin over ADP-Pi-actin [64]. However cortactin itself is not thought to nucleate actin within the lamellipodium [66], but rather serves to stabilize newly polymerized actin filaments in order to promote lamellipodial persistence [64, 66].
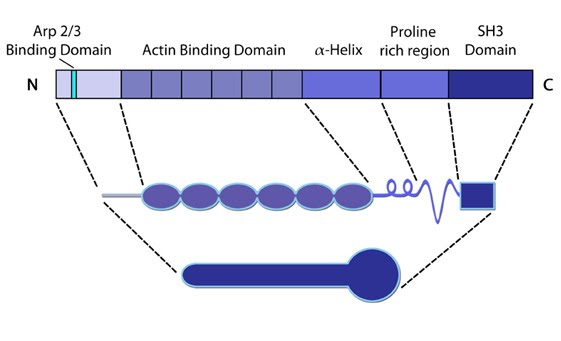 Figure 6. Cortactin: This schematic diagram illustrates the molecular organization of cortactin (reviewed in [23])
and provides examples for how cortactin is represented in figures
throughout this resource. Relevant domains believed to be important for
cortactin activity are highlighted.
Figure 6. Cortactin: This schematic diagram illustrates the molecular organization of cortactin (reviewed in [23])
and provides examples for how cortactin is represented in figures
throughout this resource. Relevant domains believed to be important for
cortactin activity are highlighted.Functionally, cortactin is involved in a wide range of cellular processes pertaining to a variety of structures. The protein is highly enriched in mature neurons and is necessary for the formation and stability of dendritic spines [62]. It also associates with post synaptic densities (PSDs) of neurons expressing the ionotropic glutamate receptor, NMDA (N-methyl-D-aspartate). This association is via a weak interaction with the PSD protein Shank [59]. The function of cortactin at PSDs is not yet clear but it is speculated to facilitate changes in the actin cytoskeleton in response to synaptic activity. This is supported by the translocation of cortactin to synapses in response to stimulation by glutamate [59].
Cortactin also forms part of the actin-rich core of both podosomes and invadopodia [63]. Actin polymerization within invadopodia is essential to the function of this structure and the migratory behavior of cancerous cells [62]. Cortactin also promotes cell motility in normal cells and as such localizes to lamellipodia [64, 65]. It enhances lamellipodial persistence i.e. increases the time spent protruding, through its interactions with both Arp2/3 and F-actin. Cortactin demonstrates a notable preference for newly polymerized actin filaments i.e. those containing a greater amount of ATP-actin over ADP-Pi-actin [64]. However cortactin itself is not thought to nucleate actin within the lamellipodium [66], but rather serves to stabilize newly polymerized actin filaments in order to promote lamellipodial persistence [64, 66].
Cortactin is also involved in non-migratory cellular processes, such as the formation of cell-cell contacts. Cortactin localizes to sites of N-cadherin mediated cell-cell adhesions, where it forms a complex with N-cadherin and serves to strengthen the intercellular contacts [67]. It also localizes to endosomal compartments [65], specifically clathrin-coated pits [68], where it is involved in receptor-mediated endocytosis. It is suggested to link the actin cytoskeleton and the endocytosing vesicle through its known interactions with F-actin, dynamin-2 and Arp2/3 [68]. These interactions are speculated to not only support a role for cortactin as a scaffold, but also to contribute to the dynamic action of endocytosis through the actin nucleating activity of cortactin [65, 68].