Cell-Adhesion[Edit]
Content
Cell adhesion is the interaction of a cell with a neighboring cell or with the underlying extracellular matrix, via specialized multi-protein adhesive structures:
Cell Matrix Adhesions
Cell Cell Adhesions
Cell Matrix Adhesions
Cell Cell Adhesions
Basic description of cell adhesion[Edit]
Cells interact with each other, and their substrate, throughout their lifetime. These interactions can be transient, such as at the immunological synapse, or they can be long-lived, such as at a neuromuscular junction. These complex cellular structures involve many proteins; from receptor molecules to structural scaffolding proteins. Significant differences in composition exist between an adhesion complex that interacts with the cellular substrate, or extracellular matrix, and one that interacts with another cell. Despite the differences however their fundamental function remains the same; to enable cellular communication through the generation and transduction of mechanical signals. While cell-cell adhesions serve as cellular ‘handshakes’, cell-matrix adhesions allow a cell to pull against its substrate to either measure the substrate rigidity, or to pull the cell forward.
Cell adhesions can be described as a functional extension of the actin cytoskeleton. Indeed, all adhesion types are linked physically to the actin filament network, and the dynamic processes of actin filament polymerization and disassembly are intertwined with the turnover and function of the adhesions complexes. Cell adhesions are mediated by either transmembrane cell-adhesion molecules (CAMs), which binding similar partner proteins on opposing cells, or adhesion receptors, which bind various ligands. These proteins are integral to the formation of adhesions and essentially link the intracellular space to the extracellular space to help relay information to the cell interior about the surroundings.
Anchoring Junctions[Edit]
Cells adhere to the ECM, or to other cells, via complexes that can collectively be called anchoring junctions (reviewed in [1, 2]). These multiprotein complexes are found in all cell types where they they stabilize the cells position, provide stability and rigidity, and support tissue integrity by holding cell sheets together. Anchoring junctions also form a tight seal between neighboring cells to restrict the flow of molecules between cells and from one side of the tissue to the other. Lastly, anchoring junctions regulate the motility of both single cells and cellular masses through their substrates. These anchor points are highly dynamic, primarily associated with actin filaments, and come in many different forms.
 Figure 1. Anchoring junctions: Actin filaments are shown in red, microtubules are yellow, and intermediate filaments are blue. Transmembrane complexes (solid ovals) contain several different components that play a key role in linking the cell exterior to the cell interior. Cell-cell adhesion: Numerous cell-cell adhesion molecules (e.g. cadherin family of proteins) and their associated cytoplasmic anchoring components (e.g. vinculin, α-actinin) form a continuum between cells and their linked cytoskeleton components. Adherens junctions (shown as a solid red oval) primarily link actin filaments between cells, while desmosomes (shown as a solid dark-blue oval) primarily link the intermediate filaments between cells. Cell-matrix adhesion: Interactions at CMACs and hemidesmosomes (shown as a solid light-blue oval) link the actin and intermediate filaments to the underlying matrix, respectively. Cell-matrix linkages are a key force-sensing unit that greatly influences cell polarity and migrationThere are three main features of anchoring junctions:
Figure 1. Anchoring junctions: Actin filaments are shown in red, microtubules are yellow, and intermediate filaments are blue. Transmembrane complexes (solid ovals) contain several different components that play a key role in linking the cell exterior to the cell interior. Cell-cell adhesion: Numerous cell-cell adhesion molecules (e.g. cadherin family of proteins) and their associated cytoplasmic anchoring components (e.g. vinculin, α-actinin) form a continuum between cells and their linked cytoskeleton components. Adherens junctions (shown as a solid red oval) primarily link actin filaments between cells, while desmosomes (shown as a solid dark-blue oval) primarily link the intermediate filaments between cells. Cell-matrix adhesion: Interactions at CMACs and hemidesmosomes (shown as a solid light-blue oval) link the actin and intermediate filaments to the underlying matrix, respectively. Cell-matrix linkages are a key force-sensing unit that greatly influences cell polarity and migrationThere are three main features of anchoring junctions:
* Transmembrane cell-adhesion molecules (CAMs) and/or adhesion receptors in the plasma membrane link the lateral surfaces of one cell to another or the basal surfaces of the cell to the ECM. Examples of adhesion receptors include cadherins (cell-cell adhesions), integrins and syndecans (cell-ECM adhesions).
* Adaptor proteins connect the adhesion molecules to the cytoskeleton and signaling molecules. Examples include catenins, talin, filamin, tensin, vinculin, and α-actinin.
* The cytoskeleton itself helps to maintain the cell shape and acts as a force-sensing device (aka mechanosensor).
There are four main types of anchoring junctions:
Several types of anchoring junctions have been identified with each involved in distinct types of adhesion.
* Adherens junctions link one cell to another cell through the actin filament network. These are found in many different cell types.
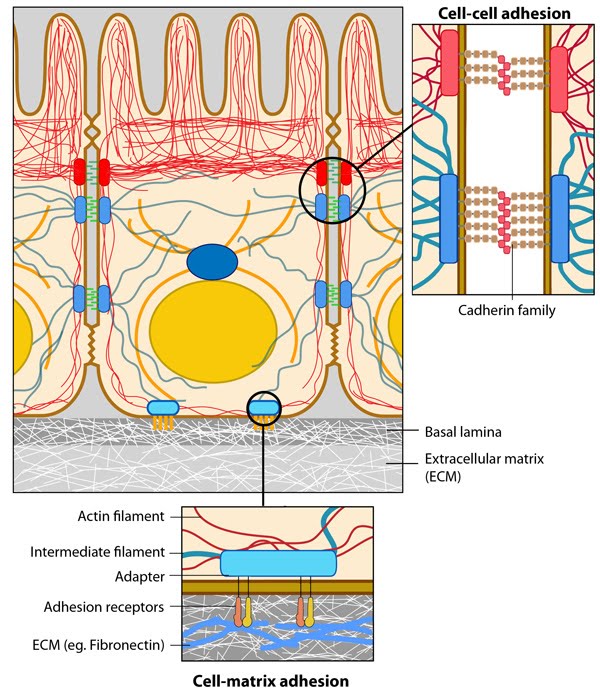 Figure 1. Anchoring junctions: Actin filaments are shown in red, microtubules are yellow, and intermediate filaments are blue. Transmembrane complexes (solid ovals) contain several different components that play a key role in linking the cell exterior to the cell interior. Cell-cell adhesion: Numerous cell-cell adhesion molecules (e.g. cadherin family of proteins) and their associated cytoplasmic anchoring components (e.g. vinculin, α-actinin) form a continuum between cells and their linked cytoskeleton components. Adherens junctions (shown as a solid red oval) primarily link actin filaments between cells, while desmosomes (shown as a solid dark-blue oval) primarily link the intermediate filaments between cells. Cell-matrix adhesion: Interactions at CMACs and hemidesmosomes (shown as a solid light-blue oval) link the actin and intermediate filaments to the underlying matrix, respectively. Cell-matrix linkages are a key force-sensing unit that greatly influences cell polarity and migration
Figure 1. Anchoring junctions: Actin filaments are shown in red, microtubules are yellow, and intermediate filaments are blue. Transmembrane complexes (solid ovals) contain several different components that play a key role in linking the cell exterior to the cell interior. Cell-cell adhesion: Numerous cell-cell adhesion molecules (e.g. cadherin family of proteins) and their associated cytoplasmic anchoring components (e.g. vinculin, α-actinin) form a continuum between cells and their linked cytoskeleton components. Adherens junctions (shown as a solid red oval) primarily link actin filaments between cells, while desmosomes (shown as a solid dark-blue oval) primarily link the intermediate filaments between cells. Cell-matrix adhesion: Interactions at CMACs and hemidesmosomes (shown as a solid light-blue oval) link the actin and intermediate filaments to the underlying matrix, respectively. Cell-matrix linkages are a key force-sensing unit that greatly influences cell polarity and migration* Transmembrane cell-adhesion molecules (CAMs) and/or adhesion receptors in the plasma membrane link the lateral surfaces of one cell to another or the basal surfaces of the cell to the ECM. Examples of adhesion receptors include cadherins (cell-cell adhesions), integrins and syndecans (cell-ECM adhesions).
* Adaptor proteins connect the adhesion molecules to the cytoskeleton and signaling molecules. Examples include catenins, talin, filamin, tensin, vinculin, and α-actinin.
* The cytoskeleton itself helps to maintain the cell shape and acts as a force-sensing device (aka mechanosensor).
There are four main types of anchoring junctions:
Types of anchoring junctions
* Adherens junctions link one cell to another cell through the actin filament network. These are found in many different cell types.
* Desmosomes link one cell to another cell through intermediate filaments. They are also found in many different types of cells.
* Hemidesmosomes link cells to the matrix through intermediate filaments. Certain hemidesmosome components also bind to F-actin (e.g. plectin [3]). Hemidesmosomes appear to be restricted to epithelial cells.
* Cell-matrix adhesion complexes (CMACs) link cells to the extracellular matrix through actin filaments. Although they are found in many different cell types, they are particularly important for regulating cell migration in motile cells.
* Hemidesmosomes link cells to the matrix through intermediate filaments. Certain hemidesmosome components also bind to F-actin (e.g. plectin [3]). Hemidesmosomes appear to be restricted to epithelial cells.
* Cell-matrix adhesion complexes (CMACs) link cells to the extracellular matrix through actin filaments. Although they are found in many different cell types, they are particularly important for regulating cell migration in motile cells.
Cell-Matrix Receptors[Edit]
Interaction between cell-matrix receptors and their respective ligands are often the initial step in the formation of a cell-matrix adhesion. Several types of cell-matrix receptors have been identified, each interacting with a specific type of ligand. Attachment of these various ECM based ligands or molecules to the exterior portion of the adhesion receptor causes their structural rearrangement [4]. This may be induced by a specific chemical property or change, (reviewed in [5]), a change in topography [6, 7, 8] (reviewed in [9, 10, 11]), or even the rigidity of the ECM components [12, 13].
 Figure 2. Integrins as adhesion receptor in focal adhesion (FA): Integrins upon binding to ECM ligand gets activated and undergoes conformational changes. This leads to a series of events including i) recruitment of binding proteins to the site leading to actin linking, ii) activation of signaling molecules leading to integrin clustering and iii) actomyosin contractions leading to adhesion strengthening. All these result in iv) formation of new actin filaments by actin polymerization modules.Cell-matrix adhesion receptors are grouped according to the ligand that is bound. In some cases, adhesion receptors bind more than one type of ligand (e.g. the integrin family) and different receptor groups may cooperate with each other to bind their ligands (reviewed in [14, 15]).Ligand-receptor binding is followed by the rapid association of other proteins to the intracellular portion of the receptor; this reinforcement of the adhesion domain is controlled by adhesion receptor mobility in the membrane [16]. Such change in forces can affect mechanosensory molecules to activate intracellular signal transduction cascades (e.g. the Rho family of GTPases) and mechanotransduction events that mediate a number of diverse processes such as cell proliferation, fate, migration, shape and polarization [17, 13] (reviewed in [18, 9, 1, 19, 20]).
Figure 2. Integrins as adhesion receptor in focal adhesion (FA): Integrins upon binding to ECM ligand gets activated and undergoes conformational changes. This leads to a series of events including i) recruitment of binding proteins to the site leading to actin linking, ii) activation of signaling molecules leading to integrin clustering and iii) actomyosin contractions leading to adhesion strengthening. All these result in iv) formation of new actin filaments by actin polymerization modules.Cell-matrix adhesion receptors are grouped according to the ligand that is bound. In some cases, adhesion receptors bind more than one type of ligand (e.g. the integrin family) and different receptor groups may cooperate with each other to bind their ligands (reviewed in [14, 15]).Ligand-receptor binding is followed by the rapid association of other proteins to the intracellular portion of the receptor; this reinforcement of the adhesion domain is controlled by adhesion receptor mobility in the membrane [16]. Such change in forces can affect mechanosensory molecules to activate intracellular signal transduction cascades (e.g. the Rho family of GTPases) and mechanotransduction events that mediate a number of diverse processes such as cell proliferation, fate, migration, shape and polarization [17, 13] (reviewed in [18, 9, 1, 19, 20]).
Various types of cell-matrix receptors exist. These include:
* Fibronectin receptors, which include the most common types of adhesion receptors, the integrin family and the syndecan family of transmembrane proteoglycans.
* Collagen receptors, which includes integrins, receptor tyrosine kinases (e.g. discoidin domain receptors [DDRs]), glycoproteins (e.g. GPVI) and immunoglobulin-like proteins aka IgCAMs (e.g. LAIR-1).
* Laminin receptors, which include integrins, the dystrophin glycoprotein complex (DGC), the 67 kDa laminin receptor (67LR), and two glycoproteins belonging to the immunoglobulin superfamily, Lutheran (Lu) and basal cell adhesion molecule (B-CAM).
* Hyaluronan receptors (aka hyaladherins) Proteins that bind to hyaluronan, a large polysaccharide, have immunoglobulin-like domains and include members of the CD44 family and CD168.
CAMs have many distinct domains that allow them to mediate cell-cell contacts by binding to specific partner proteins; when these interactions occur between apposed cells they are described as either homophilic (i.e. binding to the same kind of CAM molecule) or heterophilic (binding to a different kind of CAM molecule). Furthermore, CAMs can mediate interactions between cells of the same type (aka homotypic adhesion) or between different cell types (aka heterotypic adhesion).
CAMs are grouped into four main families:
* Cadherins: Mediate primarily homophilic interactions at cell-cell adhesions.
* Immunoglobulin superfamily (Ig) CAMs: Mediate homophilic interactions at cell-cell adhesions.
* Integrins: Some integrin types mediate heterophilic cell-cell interactions.
* Selectins: Mediate heterophilic interactions at cell-cell adhesions.
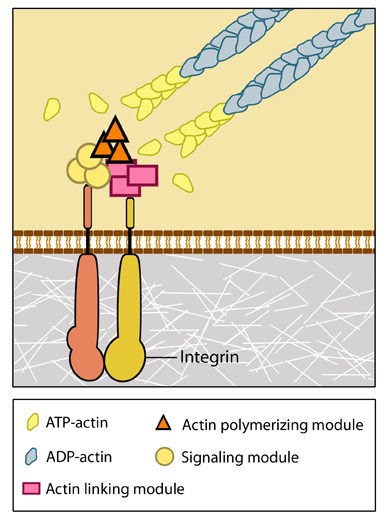 Figure 2. Integrins as adhesion receptor in focal adhesion (FA): Integrins upon binding to ECM ligand gets activated and undergoes conformational changes. This leads to a series of events including i) recruitment of binding proteins to the site leading to actin linking, ii) activation of signaling molecules leading to integrin clustering and iii) actomyosin contractions leading to adhesion strengthening. All these result in iv) formation of new actin filaments by actin polymerization modules.Cell-matrix adhesion receptors are grouped according to the ligand that is bound. In some cases, adhesion receptors bind more than one type of ligand (e.g. the integrin family) and different receptor groups may cooperate with each other to bind their ligands (reviewed in [14, 15]).
Figure 2. Integrins as adhesion receptor in focal adhesion (FA): Integrins upon binding to ECM ligand gets activated and undergoes conformational changes. This leads to a series of events including i) recruitment of binding proteins to the site leading to actin linking, ii) activation of signaling molecules leading to integrin clustering and iii) actomyosin contractions leading to adhesion strengthening. All these result in iv) formation of new actin filaments by actin polymerization modules.Cell-matrix adhesion receptors are grouped according to the ligand that is bound. In some cases, adhesion receptors bind more than one type of ligand (e.g. the integrin family) and different receptor groups may cooperate with each other to bind their ligands (reviewed in [14, 15]).Various types of cell-matrix receptors exist. These include:
* Fibronectin receptors, which include the most common types of adhesion receptors, the integrin family and the syndecan family of transmembrane proteoglycans.
* Collagen receptors, which includes integrins, receptor tyrosine kinases (e.g. discoidin domain receptors [DDRs]), glycoproteins (e.g. GPVI) and immunoglobulin-like proteins aka IgCAMs (e.g. LAIR-1).
* Laminin receptors, which include integrins, the dystrophin glycoprotein complex (DGC), the 67 kDa laminin receptor (67LR), and two glycoproteins belonging to the immunoglobulin superfamily, Lutheran (Lu) and basal cell adhesion molecule (B-CAM).
* Hyaluronan receptors (aka hyaladherins) Proteins that bind to hyaluronan, a large polysaccharide, have immunoglobulin-like domains and include members of the CD44 family and CD168.
Cell-Adhesion Molecules
Anchoring junctions are multiprotein complexes. Crucial to the formation of these junctions are cell adhesion molecules (CAMs).CAMs have many distinct domains that allow them to mediate cell-cell contacts by binding to specific partner proteins; when these interactions occur between apposed cells they are described as either homophilic (i.e. binding to the same kind of CAM molecule) or heterophilic (binding to a different kind of CAM molecule). Furthermore, CAMs can mediate interactions between cells of the same type (aka homotypic adhesion) or between different cell types (aka heterotypic adhesion).
CAMs are grouped into four main families:
* Cadherins: Mediate primarily homophilic interactions at cell-cell adhesions.
* Immunoglobulin superfamily (Ig) CAMs: Mediate homophilic interactions at cell-cell adhesions.
* Integrins: Some integrin types mediate heterophilic cell-cell interactions.
* Selectins: Mediate heterophilic interactions at cell-cell adhesions.
Cadherins[Edit]
This family of glycoproteins includes over 100 members divided into 6 subfamilies; type I classical cadherins, type II atypical cadherins, desmosomal cadherins, flamingo cadherins, proto-cadherins and several ungrouped members. Cadherins can be identified through common motifs in their extracellular domains termed cadherin repeats. Not all cadherins are involved in cell-cell adhesion, though type I and type II cadherins have well established roles in this process [21]. Both of these subfamilies contain cadherin repeats within their extracellular domains, with the outermost cadherin repeats facilitating extracellular interactions with cadherins on apposing cells (transinteractions). Type I cadherins can in addition engage in lateral interactions on the same cell (cis interactions)[22]. Intercellular interactions between cadherins can occur between those of the same type (homophilic binding) or a different type (heterophilic binding). Intracellular interactions involve the cytoplasmic domains of the cadherins. In the case of type I cadherins these interactions can be used to identify this subfamily, namely through their ability to bind catenins via their cytoplasmic tails. Catenins form part of the bridge connecting adherens junctions to the actin cytoskeleton. It should be noted that individual cadherin interactions are weak. The strength of cadherin-based adhesive junctions comes from the clustering of multiple, weak cadherin-cadherin interactions [23].
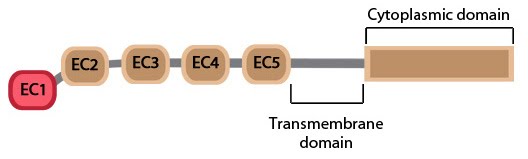
The cadherin protein family are common cell-adhesion molecules (CAMs) that mediate cell-cell contacts at anchoring junctions (e.g. adherens junctions, desmosomes) and at prominent sites of cell-cell communication (e.g. neuronal synapses). There are over 100 different cadherin family members that are grouped into at least 6 subfamilies, including type I classical cadherins, type II atypical cadherins and desmosomal cadherins [24]. All cadherins share a common architecture in their extracellular domain that comprises cadherin repeats, with classical cadherins containing five of these repeats (see Figure below). Subtle differences between cadherins impart each type with specificity for particular tissue and cell types. Cadherins use a common set of adaptor molecules and pathways to facilitate cell adhesion and communication, however the strength of adhesion varies with the type of cadherin present [25].
 Figure 4. Types of cadherin interactions: All cadherins have a common extracellular domain that is structured into tandem blocks that are variable in number and are called ‘cadherin repeats’; classical cadherins have five cadherin repeats (as shown). The cadherin intracellular domain is bound by adaptor proteins. (A) Example of a cis interaction (i.e. on the same cell) that is homophilic (i.e. cadherins of the same type). (B) Example of a trans interaction (i.e. between two cells) that is homophilic. (C) Example of a trans interaction that is heterophilic (between two different types of cadherin).
Most cadherins adhere by homophilic interactions (i.e. they bind to the same type of cadherin) but certain types (e.g. E-cadherin) also adhere by heterophilic interactions (i.e. they bind other types of cadherin). Cadherin association is sensitive to extracellular calcium (hence their name, calcium adhering). The interactions can take place laterally on the same cell, called a cis interaction, or between two cells, called a trans interaction (see Figure below).
Figure 4. Types of cadherin interactions: All cadherins have a common extracellular domain that is structured into tandem blocks that are variable in number and are called ‘cadherin repeats’; classical cadherins have five cadherin repeats (as shown). The cadherin intracellular domain is bound by adaptor proteins. (A) Example of a cis interaction (i.e. on the same cell) that is homophilic (i.e. cadherins of the same type). (B) Example of a trans interaction (i.e. between two cells) that is homophilic. (C) Example of a trans interaction that is heterophilic (between two different types of cadherin).
Most cadherins adhere by homophilic interactions (i.e. they bind to the same type of cadherin) but certain types (e.g. E-cadherin) also adhere by heterophilic interactions (i.e. they bind other types of cadherin). Cadherin association is sensitive to extracellular calcium (hence their name, calcium adhering). The interactions can take place laterally on the same cell, called a cis interaction, or between two cells, called a trans interaction (see Figure below).
Structurally, classical cadherins have five Ca+2-dependent extracellular domains and a relatively short cytoplasmic domain. Although cadherin-cadherin binding between the extracellular domains is relatively weak, the conformational changes that are induced after binding imparts the individual cadherins with rigidity. This stabilizes the interaction and fosters additional lateral cis interactions with other cadherins and generates tighter adhesions. Increased clustering of cadherins at sites of cell-cell contact correlates with increased stability and maturation of actin-based structures such as dendritic spines [4].
The classical cadherins (e.g. E-, N-, and P-cadherins) are the most common family members. Classical cadherins interact directly with p120ctn at their transmembrane region and through their cytoplasmic tails to beta (β)-catenin or plakoglobin (i.e. gamma [γ]-catenin). The correct function and stability of the cadherins requires these associations (reviewed in [26]). β-catenin binds tightly to classical cadherins before they are transported to the cell surface [27, 28]). Cadherins further interact indirectly with other adaptor proteins (e.g. alpha [α]-catenin, vinculin, EPLIN, α-actinin, zyxin) to form linkages between the cell membrane and the actin cytoskeleton (reviewed in [8]). Desmosomes in contrast, have two specialized cadherins that interact with specific adaptor proteins (e.g. plakoglobin, plakophilin, desmoplakin) to form links with the intermediate filaments.
Cadherin subfamilies:
* Type I classical cadherins – includes epithelial (E)-, neural (N)- and placental (P)-cadherin
* Type II atypical cadherins – includes vascular endothelial (VE)-cadherin
* Desmosomal cadherins – includes desmoglein and desmocollin
* Flamingo cadherins
* Proto-cadherins
The cadherin protein family are common cell-adhesion molecules (CAMs) that mediate cell-cell contacts at anchoring junctions (e.g. adherens junctions, desmosomes) and at prominent sites of cell-cell communication (e.g. neuronal synapses). There are over 100 different cadherin family members that are grouped into at least 6 subfamilies, including type I classical cadherins, type II atypical cadherins and desmosomal cadherins [24]. All cadherins share a common architecture in their extracellular domain that comprises cadherin repeats, with classical cadherins containing five of these repeats (see Figure below). Subtle differences between cadherins impart each type with specificity for particular tissue and cell types. Cadherins use a common set of adaptor molecules and pathways to facilitate cell adhesion and communication, however the strength of adhesion varies with the type of cadherin present [25].
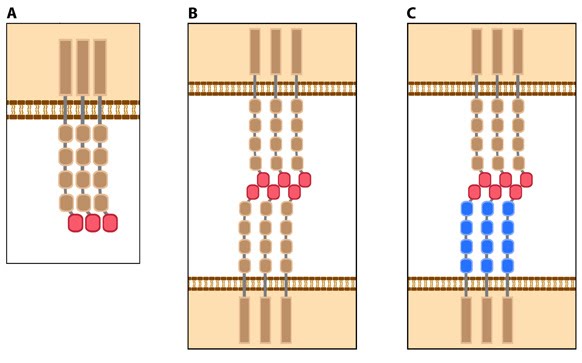 Figure 4. Types of cadherin interactions: All cadherins have a common extracellular domain that is structured into tandem blocks that are variable in number and are called ‘cadherin repeats’; classical cadherins have five cadherin repeats (as shown). The cadherin intracellular domain is bound by adaptor proteins. (A) Example of a cis interaction (i.e. on the same cell) that is homophilic (i.e. cadherins of the same type). (B) Example of a trans interaction (i.e. between two cells) that is homophilic. (C) Example of a trans interaction that is heterophilic (between two different types of cadherin).
Figure 4. Types of cadherin interactions: All cadherins have a common extracellular domain that is structured into tandem blocks that are variable in number and are called ‘cadherin repeats’; classical cadherins have five cadherin repeats (as shown). The cadherin intracellular domain is bound by adaptor proteins. (A) Example of a cis interaction (i.e. on the same cell) that is homophilic (i.e. cadherins of the same type). (B) Example of a trans interaction (i.e. between two cells) that is homophilic. (C) Example of a trans interaction that is heterophilic (between two different types of cadherin).Structurally, classical cadherins have five Ca+2-dependent extracellular domains and a relatively short cytoplasmic domain. Although cadherin-cadherin binding between the extracellular domains is relatively weak, the conformational changes that are induced after binding imparts the individual cadherins with rigidity. This stabilizes the interaction and fosters additional lateral cis interactions with other cadherins and generates tighter adhesions. Increased clustering of cadherins at sites of cell-cell contact correlates with increased stability and maturation of actin-based structures such as dendritic spines [4].
The classical cadherins (e.g. E-, N-, and P-cadherins) are the most common family members. Classical cadherins interact directly with p120ctn at their transmembrane region and through their cytoplasmic tails to beta (β)-catenin or plakoglobin (i.e. gamma [γ]-catenin). The correct function and stability of the cadherins requires these associations (reviewed in [26]). β-catenin binds tightly to classical cadherins before they are transported to the cell surface [27, 28]). Cadherins further interact indirectly with other adaptor proteins (e.g. alpha [α]-catenin, vinculin, EPLIN, α-actinin, zyxin) to form linkages between the cell membrane and the actin cytoskeleton (reviewed in [8]). Desmosomes in contrast, have two specialized cadherins that interact with specific adaptor proteins (e.g. plakoglobin, plakophilin, desmoplakin) to form links with the intermediate filaments.
Cadherin subfamilies:
* Type I classical cadherins – includes epithelial (E)-, neural (N)- and placental (P)-cadherin
* Type II atypical cadherins – includes vascular endothelial (VE)-cadherin
* Desmosomal cadherins – includes desmoglein and desmocollin
* Flamingo cadherins
* Proto-cadherins
* Ungrouped cadherins
Immunoglobulin superfamily (Ig) CAMs[Edit]
Members of this family include vascular and neural cell adhesions molecules (VCAM and NCAM), intercellular adhesion molecules (ICAM) and the nectins and nectin-like (Necl) proteins. Nectins in particular are involved in the formation of cadherin-based cell-cell junctions [29], mediating initial cell-cell contacts via nectin-nectin or nectin-Necl binding and establishing links to the actin cytoskeleton via nectin-afadin binding [30]. Of the four major groups of CAMs, IgCAMs are the only group that function independently of calcium.
The Ig superfamily is a large group of cell surface molecules that includes members such as:
 Figure 5. Schematic diagram of immunoglobulin superfamily members that are found in neurons: The extracytoplasmic portion of all Ig superfamily members is composed of a variable number of Ig domains (~3.7 nm per Ig domain) that together yield a final length of approximately 17-19 nm for both NCAM [31] and ICAM-1 [32]; certain Ig family members also contain a variable number of fibronectin III repeat domains. The transmembrane domain (TM) located at the extreme carboxy-terminus leaves a relatively small portion on the intracellular side for associating with other proteins. ICAM-1 is similar to ICAM-5 (not shown) but contains five Ig domains. Nectin and Necls share a similar structure, with three extracytoplasmic Ig-like domains (reviewed in [33])
Members of the Ig superfamily resemble each other in their three-dimensional structure as well as their amino acid sequence (reviewed in [34]). ICAMs and NCAMs form heterophilic and homophilic interactions (respectively) with adhesion molecules on other cells through a rigid extracytoplasmic rod domain that contains at least one flexible hinge domain [35, 36]. Although nectins and Necls form both heterophilic and homophilic interactions (reviewed in[29]), their homophilic interactions tend to be stronger [37].
Figure 5. Schematic diagram of immunoglobulin superfamily members that are found in neurons: The extracytoplasmic portion of all Ig superfamily members is composed of a variable number of Ig domains (~3.7 nm per Ig domain) that together yield a final length of approximately 17-19 nm for both NCAM [31] and ICAM-1 [32]; certain Ig family members also contain a variable number of fibronectin III repeat domains. The transmembrane domain (TM) located at the extreme carboxy-terminus leaves a relatively small portion on the intracellular side for associating with other proteins. ICAM-1 is similar to ICAM-5 (not shown) but contains five Ig domains. Nectin and Necls share a similar structure, with three extracytoplasmic Ig-like domains (reviewed in [33])
Members of the Ig superfamily resemble each other in their three-dimensional structure as well as their amino acid sequence (reviewed in [34]). ICAMs and NCAMs form heterophilic and homophilic interactions (respectively) with adhesion molecules on other cells through a rigid extracytoplasmic rod domain that contains at least one flexible hinge domain [35, 36]. Although nectins and Necls form both heterophilic and homophilic interactions (reviewed in[29]), their homophilic interactions tend to be stronger [37].
- vascular cell adhesion molecules (VCAM)
- neural cell adhesion molecules (NCAM)
- intercellular adhesion molecules (ICAM)
- nectin and nectin-like (Necl) family
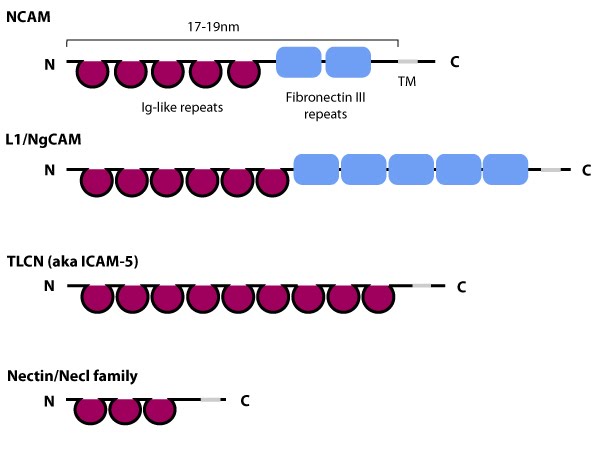 Figure 5. Schematic diagram of immunoglobulin superfamily members that are found in neurons: The extracytoplasmic portion of all Ig superfamily members is composed of a variable number of Ig domains (~3.7 nm per Ig domain) that together yield a final length of approximately 17-19 nm for both NCAM [31] and ICAM-1 [32]; certain Ig family members also contain a variable number of fibronectin III repeat domains. The transmembrane domain (TM) located at the extreme carboxy-terminus leaves a relatively small portion on the intracellular side for associating with other proteins. ICAM-1 is similar to ICAM-5 (not shown) but contains five Ig domains. Nectin and Necls share a similar structure, with three extracytoplasmic Ig-like domains (reviewed in [33])
Figure 5. Schematic diagram of immunoglobulin superfamily members that are found in neurons: The extracytoplasmic portion of all Ig superfamily members is composed of a variable number of Ig domains (~3.7 nm per Ig domain) that together yield a final length of approximately 17-19 nm for both NCAM [31] and ICAM-1 [32]; certain Ig family members also contain a variable number of fibronectin III repeat domains. The transmembrane domain (TM) located at the extreme carboxy-terminus leaves a relatively small portion on the intracellular side for associating with other proteins. ICAM-1 is similar to ICAM-5 (not shown) but contains five Ig domains. Nectin and Necls share a similar structure, with three extracytoplasmic Ig-like domains (reviewed in [33])ICAM family
Most ICAMs are expressed mainly by immune cells and endothelial cells, however brain-specific forms also exist (e.g. ICAM-5 aka TLCN) [38, 39]. All ICAMs appear to share lymphocyte function-associated antigen-1 (LFA-1, CD11a/CD18, αLβ2 integrin) as their counter receptor [40, 41, 42]. LFA-1 integrin is found on the surface of leukocytes where it modulates adhesion-dependent events that are essential for immune system activities. In the brain, LFA-1 expression appears to be restricted to resident macrophages (microglia) and its expression is tied to microglia activation [43]. ICAM-1 and LFA-1 binding is magnesium-dependent [40, 44] and the sites for LFA-1 binding lie in the first two amino-terminal Ig domains of ICAM-1; the residues involved in binding to LFA-1 are conserved in other ICAMs [35].Nectin and Necl family
Nectins and nectin-like molecules (Necls) are expressed in a number of cell types where they have been shown to be important for cell-cell adhesion and the formation of stable junctions (e.g adherens junctions). Nectins and Necls also play a role in various cellular activities including cell polarization, migration, growth and cell fate (reviewed in [29, 45]). Nectin and Necls interact with and share a number of binding partners through their cytoplasmic domain, however, only nectins bind to afadin, an F-actin binding protein.α-catenin[Edit]
α-catenin has broad activity that contributes to processes such as differentiation (i.e. commitment to a particular cell type), embryonic and tissue development, and cell migration (reviewed in [46, 47]). α-catenin is concentrated at cell-cell adhesion sites, e.g., tight junctions [48], and adherens junctions (reviewed in [2]), through its association with a related family member, beta (β)-catenin; this binding interaction is controlled by phosphorylation of either α- or β-catenin [49, 50] (reviewed in [51]) and phosphorylated β-catenin is expected to compete with homodimerziation of α-catenin [49, 52]. Dimerization of α-catenin creates a complex with functional domains at both ends that preferentially binds actin filaments, in contrast to the monomer which prefers E-cadherin-β-catenin complexes [53].
 Figure 6. Alpha (α)-catenin: This schematic diagram illustrates the molecular organization of
α-catenin and provides examples for how α-catenin is represented in
figures throughout this resource.Because β-catenin binds tightly to classical cadherins before they are transported to the cell surface [27, 28]), it was originally suggested that α-catenin formed an indirect link between adhesion receptors and the actin cytoskeleton via its association with β-catenin or other actin-binding proteins (e.g. vinculin and α-actinin) [28]. However, later work showed that α-catenin cannot bind the E-cadherin-β-catenin complex and actin simultaneously, nor does it bind actin indirectly through its binding partners, vinculin or α-actinin [54]. Thus, rather than serving as a bridge to the cytoskeleton, α-catenin appears to function primarily as a molecular switch that promotes stable cell-cell adhesions (e.g. adherens junctions).
Figure 6. Alpha (α)-catenin: This schematic diagram illustrates the molecular organization of
α-catenin and provides examples for how α-catenin is represented in
figures throughout this resource.Because β-catenin binds tightly to classical cadherins before they are transported to the cell surface [27, 28]), it was originally suggested that α-catenin formed an indirect link between adhesion receptors and the actin cytoskeleton via its association with β-catenin or other actin-binding proteins (e.g. vinculin and α-actinin) [28]. However, later work showed that α-catenin cannot bind the E-cadherin-β-catenin complex and actin simultaneously, nor does it bind actin indirectly through its binding partners, vinculin or α-actinin [54]. Thus, rather than serving as a bridge to the cytoskeleton, α-catenin appears to function primarily as a molecular switch that promotes stable cell-cell adhesions (e.g. adherens junctions).
Current models suggest that α-catenin promotes stronger adhesions in a few ways: 1) α-catenin may foster lateral clustering and activation of cadherins [55]; 2) α-catenin may recruit formins at nascent cell-cell contacts to produce new filaments that push against and bring the membranes together [56] (reviewed in [46, 47]); and 3) α-catenin may regulate membrane protrusive activity [57] and suppress Arp2/3 complex-mediated actin nucleation and polymerization at the leading edge in the lamellipodium [53] (reviewed in [47]).
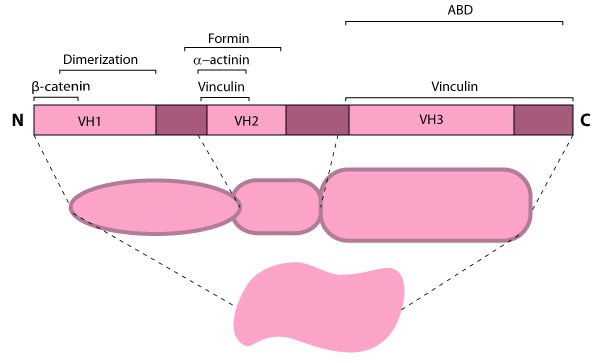 Figure 6. Alpha (α)-catenin: This schematic diagram illustrates the molecular organization of
α-catenin and provides examples for how α-catenin is represented in
figures throughout this resource.
Figure 6. Alpha (α)-catenin: This schematic diagram illustrates the molecular organization of
α-catenin and provides examples for how α-catenin is represented in
figures throughout this resource.Current models suggest that α-catenin promotes stronger adhesions in a few ways: 1) α-catenin may foster lateral clustering and activation of cadherins [55]; 2) α-catenin may recruit formins at nascent cell-cell contacts to produce new filaments that push against and bring the membranes together [56] (reviewed in [46, 47]); and 3) α-catenin may regulate membrane protrusive activity [57] and suppress Arp2/3 complex-mediated actin nucleation and polymerization at the leading edge in the lamellipodium [53] (reviewed in [47]).
Integrins and Selectins[Edit]
Integrins
These CAMs form heterodimers comprising an alpha and beta subunit and are commonly known to facilitate cell-matrix interactions (e.g. at focal adhesions) via their interactions with extracellular matrix proteins. However they are also capable of mediating cell-cell interactions through their interactions with IgCAMs – a process vital in mounting immune responses via leukocytes [58].Selectins
Three members constitute this family, E-selectin (endothelial), L-selectin (leukocyte) and P-selectin (platelet), all of which bind to fucosylated carbohydrates [59]. For example P-selectin on leukocytes binds PSGL-1 (P-selectin glycoprotein ligand-1) on endothelial cells.Whether an adhesion is formed between two cells, or between a cell and its substrate, alterations to the actin cytoskeleton occur. This is because the adhesion complex must connect to the cytoskeleton in order facilitate its function. Several proteins help facilitate this process including Ena/VASP which associates with components of the Arp2/3-mediated actin assembly module. These are required for actin dynamics at sites of cadherin-cadherin binding [60]. The association of Mena and VASP may be modulated by signal-mediated phosphorylation (reviewed in [61]); VASP phosphorylation prevents it from interacting with other cadherin-complex proteins (e.g. zyxin) [62].
Parvin[Edit]
Adaptor proteins Other proteins located at sites of cell adhesion include the adaptor proteins which connect the adhesion molecules to the cytoskeleton and signaling molecules. Examples include parvin, paxillin, talin, tensin, vinculin and zyxin.
 Figure 7. Parvin: This schematic diagram illustrates the molecular organization of parvin and provides examples for how parvin is presented in figures throughout this resource. Relevant domains believed to be important for binding to actin (e.g. CH1, CH2) and protein-protein interactions are highlighted (reviewed in [63])Although CH domains are a common feature shared between members of the α-actinin superfamily (reviewed in [65]), the ABDs of parvin are unique and more closely related to fimbrin, which further separates the parvins into a separate family within the α-actinin superfamily [64]. Furthermore, the CH domains of parvin were suggested to have evolved for specifically interacting with non-actin targets at sites of focal adhesion assembly; for example, recruitment of parvin to focal adhesions (FAs) requires an association with paxillin via a paxillin-binding sequence (PBS) motif contained within second CH domain of parvin [66]. Similar to other members of the α-actinin superfamily, α-parvin (aka actopaxin) may function as a dimer [67].
Figure 7. Parvin: This schematic diagram illustrates the molecular organization of parvin and provides examples for how parvin is presented in figures throughout this resource. Relevant domains believed to be important for binding to actin (e.g. CH1, CH2) and protein-protein interactions are highlighted (reviewed in [63])Although CH domains are a common feature shared between members of the α-actinin superfamily (reviewed in [65]), the ABDs of parvin are unique and more closely related to fimbrin, which further separates the parvins into a separate family within the α-actinin superfamily [64]. Furthermore, the CH domains of parvin were suggested to have evolved for specifically interacting with non-actin targets at sites of focal adhesion assembly; for example, recruitment of parvin to focal adhesions (FAs) requires an association with paxillin via a paxillin-binding sequence (PBS) motif contained within second CH domain of parvin [66]. Similar to other members of the α-actinin superfamily, α-parvin (aka actopaxin) may function as a dimer [67].
Parvin
The parvins are a family of actin binding proteins (known as α-, β- and γ-parvin in mammals) that are members of the actin linking functional module at cell-matrix adhesion sites (reviewed in [63]). Parvin is a small protein (42 kDa) that contains a variable amino terminus followed by two actin-binding domains (ABDs) each composed from two calponin homology (CH) domains [64].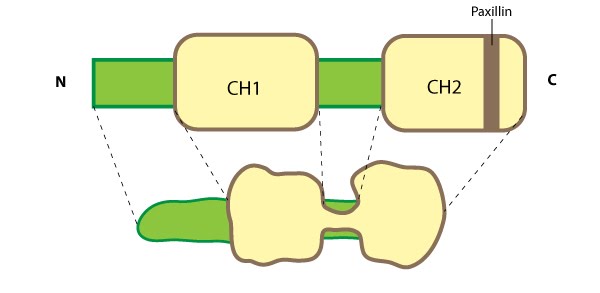 Figure 7. Parvin: This schematic diagram illustrates the molecular organization of parvin and provides examples for how parvin is presented in figures throughout this resource. Relevant domains believed to be important for binding to actin (e.g. CH1, CH2) and protein-protein interactions are highlighted (reviewed in [63])
Figure 7. Parvin: This schematic diagram illustrates the molecular organization of parvin and provides examples for how parvin is presented in figures throughout this resource. Relevant domains believed to be important for binding to actin (e.g. CH1, CH2) and protein-protein interactions are highlighted (reviewed in [63])Parvin is found in several cell types and at many locations in the cell such as: the leading edge of migrating cells and at sites of growing adhesions; it extends from mature FAs; and it partially localizes with stress fibers [64, 66]. Parvin co-localizes completely with talin in FAs and with fibers along the cell body [64]. As parvin is not usually found along the entire length of stress fibers [66], these central fibers more likely resemble tensin-rich fibrillar adhesions (FBs) [68]. Parvin is a member of a triad known as IPP (ILK-PINCH-parvin) which controls the maturation of cell-matrix adhesions by forming a permissive platform for tensin recruitment [69]. Parvin contains numerous potential phosphorylation consensus sequences for kinases such as protein kinase C [66] and extracellular signal-regulated protein kinase [70]; phosphoryation of parvin increases during cell adhesion/spreading [70].
Paxillin[Edit]
Paxillin is a multidomain scaffolding protein that is a key platform for bringing together signaling molecules, structural components, and regulatory proteins that control the adhesion and organization of the internal cytoskeleton for processes such as cell migration (reviewed in [71]).  Figure 8. Paxillin: This schematic diagram illustrates the molecular organization of paxillin and provides examples for how paxillin is represented in figures throughout this resource. Relevant domains believed to be important for protein-protein interactions are highlighted (reviewed in [72]). These interactions include FAK, vinculin [73, 74], Src [75], parvin [76],tubulin [77], α-integrin [78, 79, 80], and focal adhesion targeting [81].Paxillin contains five amino-terminal leucine-aspartic acid (LD1-5) motifs and four carboxy-terminal LIM (Lin11, Isl-1, Mec-3) domains; the LD and LIM domains mediate protein-protein interactions with a number of structural and regulatory proteins (see figure at right).
Figure 8. Paxillin: This schematic diagram illustrates the molecular organization of paxillin and provides examples for how paxillin is represented in figures throughout this resource. Relevant domains believed to be important for protein-protein interactions are highlighted (reviewed in [72]). These interactions include FAK, vinculin [73, 74], Src [75], parvin [76],tubulin [77], α-integrin [78, 79, 80], and focal adhesion targeting [81].Paxillin contains five amino-terminal leucine-aspartic acid (LD1-5) motifs and four carboxy-terminal LIM (Lin11, Isl-1, Mec-3) domains; the LD and LIM domains mediate protein-protein interactions with a number of structural and regulatory proteins (see figure at right).
Paxillin contains a number of likely phosphorylation sites for serine/threonine kinases (e.g. protein kinase C) and tyrosine kinases [82] and is phosphorylated in response to various growth factors and adhesion stimuli both in vitro [83, 84] and in vivo [85](reviewed in [71]). Phosphorylation of the LIM domains has been suggested to influence cellular adhesion to fibronectin as well as paxillin localization to focal adhesions [86].
 Figure 9. Paxillin Localization: A HFF (human foreskin fibroblast) cell plated on a fibronectin coated glass coverslip, transfected with GFP-LifeAct (green), which labels F-actin in living cells, and mCherry-paxillin (red). It was imaged on Olympus IX81 Inverted microscope using a Perkin Elmer spinning disk at 100x magnification. Image courtesy: Yee Han Tee, Mechanobiology Institute, Singapore.Paxillin binds directly to α-integrins via its amino terminus [87, 88, 89] and it localizes specifically to sites of cell-matrix adhesion (as opposed to cell-cell contacts) (see microphotograph below) [82]. Paxillin also co-localizes with talin and vinculin at the ends of stress fibers [82]. Mechanical tension and force from actin/myosin based contractions along the cytoskeleton network are necessary not only for paxillin recruitment at the adhesion sites during their maturation [90] as well as to stabilize and maintain its localization [91, 92]. These findings, together with evidence suggesting the involvement of paxillin in the detection of shear stress [93], makes paxillin a likely candidate for a mechanosensor.
Figure 9. Paxillin Localization: A HFF (human foreskin fibroblast) cell plated on a fibronectin coated glass coverslip, transfected with GFP-LifeAct (green), which labels F-actin in living cells, and mCherry-paxillin (red). It was imaged on Olympus IX81 Inverted microscope using a Perkin Elmer spinning disk at 100x magnification. Image courtesy: Yee Han Tee, Mechanobiology Institute, Singapore.Paxillin binds directly to α-integrins via its amino terminus [87, 88, 89] and it localizes specifically to sites of cell-matrix adhesion (as opposed to cell-cell contacts) (see microphotograph below) [82]. Paxillin also co-localizes with talin and vinculin at the ends of stress fibers [82]. Mechanical tension and force from actin/myosin based contractions along the cytoskeleton network are necessary not only for paxillin recruitment at the adhesion sites during their maturation [90] as well as to stabilize and maintain its localization [91, 92]. These findings, together with evidence suggesting the involvement of paxillin in the detection of shear stress [93], makes paxillin a likely candidate for a mechanosensor.
In migrating cells, paxillin appears to remodel from older to newer adhesions at the leading edge to become one of the first proteins found at cell-matrix adhesion sites [94]. Paxillin largely contributes to cytoskeleton dynamics by regulating the activity of the Rho family of GTPases and by coordinating their association with specific ligands and downstream effector systems [71]; for example, paxillin-integrin binding is sufficient for regulating signal transduction through Rac1 GTPase [89]. It has also been recently shown to coordinate membrane trafficking and hence directional migration based on physical cues [95].
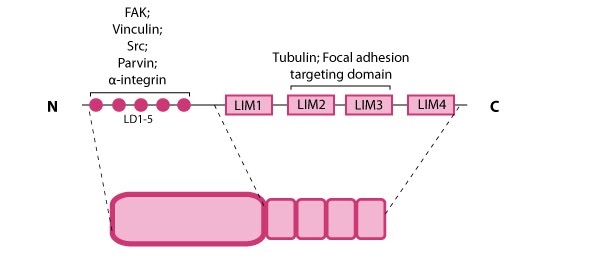 Figure 8. Paxillin: This schematic diagram illustrates the molecular organization of paxillin and provides examples for how paxillin is represented in figures throughout this resource. Relevant domains believed to be important for protein-protein interactions are highlighted (reviewed in [72]). These interactions include FAK, vinculin [73, 74], Src [75], parvin [76],tubulin [77], α-integrin [78, 79, 80], and focal adhesion targeting [81].
Figure 8. Paxillin: This schematic diagram illustrates the molecular organization of paxillin and provides examples for how paxillin is represented in figures throughout this resource. Relevant domains believed to be important for protein-protein interactions are highlighted (reviewed in [72]). These interactions include FAK, vinculin [73, 74], Src [75], parvin [76],tubulin [77], α-integrin [78, 79, 80], and focal adhesion targeting [81].Paxillin contains a number of likely phosphorylation sites for serine/threonine kinases (e.g. protein kinase C) and tyrosine kinases [82] and is phosphorylated in response to various growth factors and adhesion stimuli both in vitro [83, 84] and in vivo [85](reviewed in [71]). Phosphorylation of the LIM domains has been suggested to influence cellular adhesion to fibronectin as well as paxillin localization to focal adhesions [86].
Localization and function
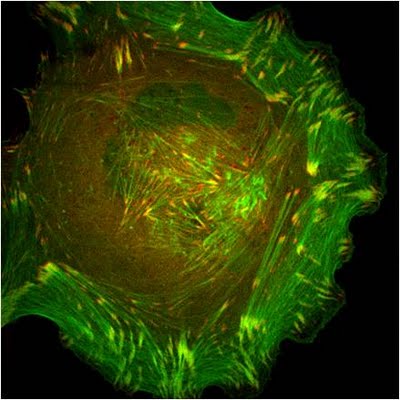 Figure 9. Paxillin Localization: A HFF (human foreskin fibroblast) cell plated on a fibronectin coated glass coverslip, transfected with GFP-LifeAct (green), which labels F-actin in living cells, and mCherry-paxillin (red). It was imaged on Olympus IX81 Inverted microscope using a Perkin Elmer spinning disk at 100x magnification. Image courtesy: Yee Han Tee, Mechanobiology Institute, Singapore.
Figure 9. Paxillin Localization: A HFF (human foreskin fibroblast) cell plated on a fibronectin coated glass coverslip, transfected with GFP-LifeAct (green), which labels F-actin in living cells, and mCherry-paxillin (red). It was imaged on Olympus IX81 Inverted microscope using a Perkin Elmer spinning disk at 100x magnification. Image courtesy: Yee Han Tee, Mechanobiology Institute, Singapore.In migrating cells, paxillin appears to remodel from older to newer adhesions at the leading edge to become one of the first proteins found at cell-matrix adhesion sites [94]. Paxillin largely contributes to cytoskeleton dynamics by regulating the activity of the Rho family of GTPases and by coordinating their association with specific ligands and downstream effector systems [71]; for example, paxillin-integrin binding is sufficient for regulating signal transduction through Rac1 GTPase [89]. It has also been recently shown to coordinate membrane trafficking and hence directional migration based on physical cues [95].
Talin[Edit]
Talin contains a 47-kDa N-terminal head, a neck and a 220kDa rod domain. The head domain comprises four subdomains termed F0, F1, F2 and F3, with the latter three forming a three-lobed FERM domain.
 Figure 10. Talin structure: This schematic diagram illustrates the molecular organization of talin (reviewed in [96, 72, 97]) and shows how talin is represented throughout this resource. Hypothetical dimer complexes are presented [98]. ABD = actin binding domain, IBS2 = integrin binding site 2, PIP2 = phosphatidylinositol-4,5-bisphosphate, FAK = focal adhesion kinase, PIPK = phosphatidylinositol phosphate type 1γ.Integrin tail binding occurs via the F3 phosphotyrosine binding (PTB) domain via a unique interaction with the integrin membrane proximal region, which is sufficient for integrin activation [99]. The basic patches on all subdomains can dock onto the plasma membrane and further enhance integrin activation. Specific interactions through basic residues on F3 are also essential for integrin clustering [100].
Figure 10. Talin structure: This schematic diagram illustrates the molecular organization of talin (reviewed in [96, 72, 97]) and shows how talin is represented throughout this resource. Hypothetical dimer complexes are presented [98]. ABD = actin binding domain, IBS2 = integrin binding site 2, PIP2 = phosphatidylinositol-4,5-bisphosphate, FAK = focal adhesion kinase, PIPK = phosphatidylinositol phosphate type 1γ.Integrin tail binding occurs via the F3 phosphotyrosine binding (PTB) domain via a unique interaction with the integrin membrane proximal region, which is sufficient for integrin activation [99]. The basic patches on all subdomains can dock onto the plasma membrane and further enhance integrin activation. Specific interactions through basic residues on F3 are also essential for integrin clustering [100].
Both F2 and F3 contribute to actin binding, with the F3 binding pocket being the same that binds integrin and PIPKIγ90 as well thus linking these adhesion components [101]. F3 also binds to layilin (a hyaluronan receptor) and signaling molecules FAK (reviewed in [102]). The neck region is susceptible to cleavage by calpain 2 [103].
The rod contains an additional integrin-binding site (IBS2), two actin-binding sites (ABD) and several vinculin-binding sites that are shown to be exposed by stretch in response to force, both in vitro [104, 105, 106] and in vivo [107]. Vinculin binding reinforces and increases the stability of adhesion sites [108]. Talin also contains numerous potential phosphorylation sites [109] which are suggested to directly or indirectly regulate the association of talin with other factors (reviewed in [102]).
 Figure 11. Talin recruitment to membrane: Talin recruitment to membrane. Ligand occupancy in certain cell-surface receptors (agonists) causes phospholipid hydrolysis releasing diacylglycerol (DAG) and inositol triphosphate (IP3). IP3 increases cytosolic levels of calcium ions; DAG and Ca2+ can promote GTP-loading and membrane translocation of Rap1 either by activating Ca2+ and DAG-regulated GEF (CALDAG-GEF or Rap-GEF) or protein kinase C (PKC). Activated Rap1 in turn, recruits Rap1-GTP-interacting adaptor molecule (RIAM) along with its binding partner, talin to the plasma membrane. Adapted from [110, 111].
Talin exists in an autoinhibited form in the cytosol due to the intermolecular association between the F3 subdomain and a helical bundle in the rod region [112, 113]. This not only blocks integrin binding site on F3 but also F2 and F3 binding to membrane. Activation predominantly occurs inside FAs [114, 115] and likely involves binding to membrane phospholipids such as phosphatidylinositol 4,5-bis-phosphate (PIP2) [116, 117] (reviewed in [118]), vinculin and F-actin [114] or calpain cleavage [103]. This enhances talin’s affinity for the β-integrin subunit by revealing binding sites.
Figure 11. Talin recruitment to membrane: Talin recruitment to membrane. Ligand occupancy in certain cell-surface receptors (agonists) causes phospholipid hydrolysis releasing diacylglycerol (DAG) and inositol triphosphate (IP3). IP3 increases cytosolic levels of calcium ions; DAG and Ca2+ can promote GTP-loading and membrane translocation of Rap1 either by activating Ca2+ and DAG-regulated GEF (CALDAG-GEF or Rap-GEF) or protein kinase C (PKC). Activated Rap1 in turn, recruits Rap1-GTP-interacting adaptor molecule (RIAM) along with its binding partner, talin to the plasma membrane. Adapted from [110, 111].
Talin exists in an autoinhibited form in the cytosol due to the intermolecular association between the F3 subdomain and a helical bundle in the rod region [112, 113]. This not only blocks integrin binding site on F3 but also F2 and F3 binding to membrane. Activation predominantly occurs inside FAs [114, 115] and likely involves binding to membrane phospholipids such as phosphatidylinositol 4,5-bis-phosphate (PIP2) [116, 117] (reviewed in [118]), vinculin and F-actin [114] or calpain cleavage [103]. This enhances talin’s affinity for the β-integrin subunit by revealing binding sites.
Agonist stimulation has been shown to trigger a signaling pathway for membrane targeting of talin/ activation of integrin αIIbβ3 [110], involving small GTPase Rap1, Rap-GEF or protein kinase C and adaptor protein RIAM [119, 120].
During cell spreading, talin undergoes cycles of stretching and vinculin binding due to contractile forces on the rearward moving actin filaments [16]. This phenomenon serves to convert matrix forces into biochemical signals at the adhesion site. Hence it not only organizes and stabilizes these initial linkages [108], but it also mediates signal transduction events through the integrins, vinculin and actin (reviewed in [102, 118, 124]).
The proteolytic cleavage of talin has been shown to be a critical event in the subsequent disassembly of other focal adhesion components [125] but not in integrin activation. Although talin is a key factor that translates mechanical forces into chemical responses primarily at sites of cell-matrix and cell-cell junctions, talin may also function in other cellular processes including membrane ruffling, cytokinesis, and phagocytosis (reviewed in [102]).
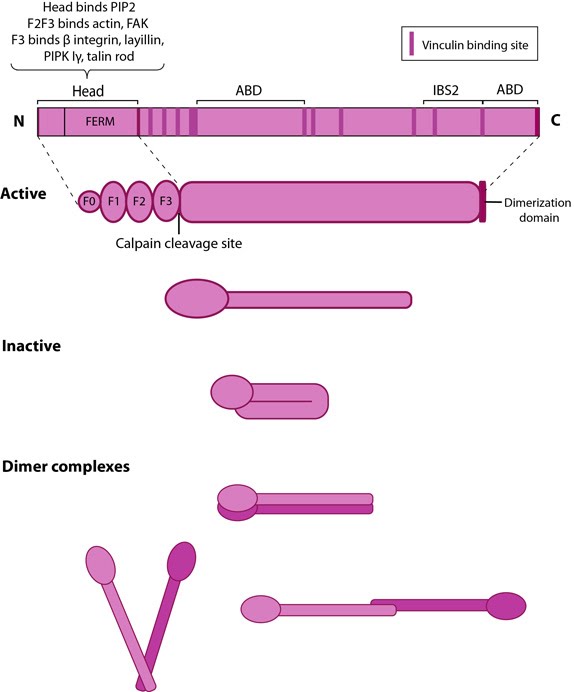 Figure 10. Talin structure: This schematic diagram illustrates the molecular organization of talin (reviewed in [96, 72, 97]) and shows how talin is represented throughout this resource. Hypothetical dimer complexes are presented [98]. ABD = actin binding domain, IBS2 = integrin binding site 2, PIP2 = phosphatidylinositol-4,5-bisphosphate, FAK = focal adhesion kinase, PIPK = phosphatidylinositol phosphate type 1γ.
Figure 10. Talin structure: This schematic diagram illustrates the molecular organization of talin (reviewed in [96, 72, 97]) and shows how talin is represented throughout this resource. Hypothetical dimer complexes are presented [98]. ABD = actin binding domain, IBS2 = integrin binding site 2, PIP2 = phosphatidylinositol-4,5-bisphosphate, FAK = focal adhesion kinase, PIPK = phosphatidylinositol phosphate type 1γ.Both F2 and F3 contribute to actin binding, with the F3 binding pocket being the same that binds integrin and PIPKIγ90 as well thus linking these adhesion components [101]. F3 also binds to layilin (a hyaluronan receptor) and signaling molecules FAK (reviewed in [102]). The neck region is susceptible to cleavage by calpain 2 [103].
The rod contains an additional integrin-binding site (IBS2), two actin-binding sites (ABD) and several vinculin-binding sites that are shown to be exposed by stretch in response to force, both in vitro [104, 105, 106] and in vivo [107]. Vinculin binding reinforces and increases the stability of adhesion sites [108]. Talin also contains numerous potential phosphorylation sites [109] which are suggested to directly or indirectly regulate the association of talin with other factors (reviewed in [102]).
Talin activation and membrane recruitment
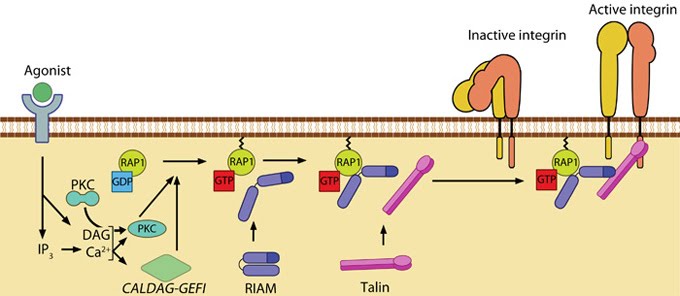 Figure 11. Talin recruitment to membrane: Talin recruitment to membrane. Ligand occupancy in certain cell-surface receptors (agonists) causes phospholipid hydrolysis releasing diacylglycerol (DAG) and inositol triphosphate (IP3). IP3 increases cytosolic levels of calcium ions; DAG and Ca2+ can promote GTP-loading and membrane translocation of Rap1 either by activating Ca2+ and DAG-regulated GEF (CALDAG-GEF or Rap-GEF) or protein kinase C (PKC). Activated Rap1 in turn, recruits Rap1-GTP-interacting adaptor molecule (RIAM) along with its binding partner, talin to the plasma membrane. Adapted from [110, 111].
Figure 11. Talin recruitment to membrane: Talin recruitment to membrane. Ligand occupancy in certain cell-surface receptors (agonists) causes phospholipid hydrolysis releasing diacylglycerol (DAG) and inositol triphosphate (IP3). IP3 increases cytosolic levels of calcium ions; DAG and Ca2+ can promote GTP-loading and membrane translocation of Rap1 either by activating Ca2+ and DAG-regulated GEF (CALDAG-GEF or Rap-GEF) or protein kinase C (PKC). Activated Rap1 in turn, recruits Rap1-GTP-interacting adaptor molecule (RIAM) along with its binding partner, talin to the plasma membrane. Adapted from [110, 111].Agonist stimulation has been shown to trigger a signaling pathway for membrane targeting of talin/ activation of integrin αIIbβ3 [110], involving small GTPase Rap1, Rap-GEF or protein kinase C and adaptor protein RIAM [119, 120].
Localization and function
Talin is abundant specifically at sites of cell-ECM linkage [121] where it appears to be a key endpoint for multiple signaling pathways that lead to integrin activation (reviewed in [122]). Talin behaves as a prominent structural platform that is required for the initial linkage between the contractile cytoskeleton and sites of integrin/fibronectin adhesion [123].During cell spreading, talin undergoes cycles of stretching and vinculin binding due to contractile forces on the rearward moving actin filaments [16]. This phenomenon serves to convert matrix forces into biochemical signals at the adhesion site. Hence it not only organizes and stabilizes these initial linkages [108], but it also mediates signal transduction events through the integrins, vinculin and actin (reviewed in [102, 118, 124]).
The proteolytic cleavage of talin has been shown to be a critical event in the subsequent disassembly of other focal adhesion components [125] but not in integrin activation. Although talin is a key factor that translates mechanical forces into chemical responses primarily at sites of cell-matrix and cell-cell junctions, talin may also function in other cellular processes including membrane ruffling, cytokinesis, and phagocytosis (reviewed in [102]).
Tensin[Edit]
Tensin is a cytoskeleton scaffolding protein that was named for its ability to form a bridge that maintains tension between the actin filaments and cell-matrix adhesion sites (reviewed in [126]).
 Figure 12. Tensin: This
schematic diagram illustrates the molecular organization of tensin and
provides examples for how tensin is represented in figures throughout
this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [126, 127]). PIP2- phosphatidylinositol [33, 128]-bis-phosphate; YP- phosphotyrosine; SH2- Src homology 2.
Figure 12. Tensin: This
schematic diagram illustrates the molecular organization of tensin and
provides examples for how tensin is represented in figures throughout
this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [126, 127]). PIP2- phosphatidylinositol [33, 128]-bis-phosphate; YP- phosphotyrosine; SH2- Src homology 2.
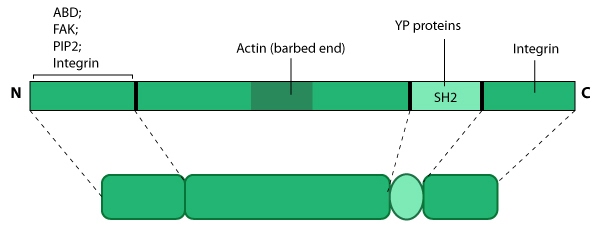 Figure 12. Tensin: This
schematic diagram illustrates the molecular organization of tensin and
provides examples for how tensin is represented in figures throughout
this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [126, 127]). PIP2- phosphatidylinositol [33, 128]-bis-phosphate; YP- phosphotyrosine; SH2- Src homology 2.
Figure 12. Tensin: This
schematic diagram illustrates the molecular organization of tensin and
provides examples for how tensin is represented in figures throughout
this resource. Relevant domains believed to be important for binding to actin and for protein-protein interactions are highlighted (reviewed in [126, 127]). PIP2- phosphatidylinositol [33, 128]-bis-phosphate; YP- phosphotyrosine; SH2- Src homology 2.Tensin contains three actin-binding domains (ABDs) that allows it to form crosslinks along actin filaments; it also prevents actin assembly by capping actin filaments at the barbed end [129, 130]. Tensin has numerous phosphorylation sites and multiple protein interaction domains for both structural components (e.g. paxillin, β-integrin [131]) and signaling molecules (e.g. Src, phosphatidylinositol 3-kinase [PI3K], focal adhesion kinase [FAK]) [132] (reviewed in [127]). Phosphorlyation of tensin corresponds with cell-ECM binding [133] and growth factor stimulation [134] (reviewed in [126]). Tensin forms a C-shaped structure [135] that binds focal adhesion components at both ends [136]. Tensin is also proposed to form a dimer via its carboxy-terminus and this association may be dependent upon its phosphorylation state [135].
Protein localization and function
Tensin primarily localizes to sites of cell attachment such as focal adhesions [136, 137], elongated fibrillar structures (aka fibrillar adhesions)[91] and possibly other adhesive junctions [138]. Tensin serves as a link between signal transduction pathways and the actin cytoskeleton by forming a structural platform that regulates the assembly of focal adhesion components, phosphoproteins, and signaling molecules for processes such as cell migration [136, 139] and tissue regeneration [140].Vinculin[Edit]
Vinculin is a protein that couples, transmits, transduces, and regulates mechanical force between the cytoskeleton and adhesion receptors (reviewed in [141]).
 Figure 13. Vinculin: This schematic diagram illustrates the molecular organization of vinculin and shows how vinculin is represented throughout this resource. Relevant domains believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [142]).Vinculin frequently links adhesion receptors (e.g. integrins) to the contractile actin–myosin cytoskeleton by binding either talin through its amino-terminal globular head domain [143], or paxillin through its rod-like tail domain [82]. Vinculin can also bind to lipids through the tail domain. The head and tail domains are linked by a flexible hinge that also contains binding sites for components of the actin polymerizing module (e.g. Arp2/3 complex [144], Ena/VASP proteins [145, 146]).
Figure 13. Vinculin: This schematic diagram illustrates the molecular organization of vinculin and shows how vinculin is represented throughout this resource. Relevant domains believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [142]).Vinculin frequently links adhesion receptors (e.g. integrins) to the contractile actin–myosin cytoskeleton by binding either talin through its amino-terminal globular head domain [143], or paxillin through its rod-like tail domain [82]. Vinculin can also bind to lipids through the tail domain. The head and tail domains are linked by a flexible hinge that also contains binding sites for components of the actin polymerizing module (e.g. Arp2/3 complex [144], Ena/VASP proteins [145, 146]).
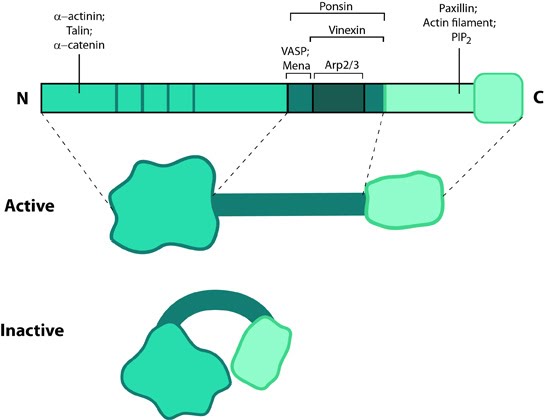 Figure 13. Vinculin: This schematic diagram illustrates the molecular organization of vinculin and shows how vinculin is represented throughout this resource. Relevant domains believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [142]).
Figure 13. Vinculin: This schematic diagram illustrates the molecular organization of vinculin and shows how vinculin is represented throughout this resource. Relevant domains believed to be important for actin binding and protein-protein interactions are highlighted (reviewed in [142]).Activation
Vinculin generally forms two structural states, an open (active) and closed (inactive) state, which are controlled by interaction(s) between the head and tail domains [147, 148]. Whether vinculin can bind to other factors depends both allosterically and sterically on the formation of the complete open state (reviewed in [142]). This in turn is favored by combinatorial binding of ligands namely talin, phosphatidylinositol 4,5-bis-phosphate [PIP2] and actin [148, 149, 150, 151](reviewed in [141]). Phosphorylation at 4 residues has been proposed to prime vinculin for this complex formation and hence the activation process [152]. Activation of vinculin influences its ability to form oligomers or other complxes in cells [153, 154].Localization
later work showed it was present in all adherens junctions [121]. Lipid binding regulates its placement in the adhesions as well as disassembly hence stimulating motility (reviewed in [142]).Vinculin recruitment to adhesion sites is mechanically regulated by ligand binding(e.g. other cells, extracellullar matrix) and activation of adhesion receptors. Physical restructuring of adhesion receptors such as integrins and their linked mechanosensors (e.g. talin) is transmitted to the cell interior to stimulate contraction of actin stress fibers; this promotes vinculin binding to these sites [143] and subsequent ordering of vinculin domains (reviewed in [148]). Interestingly, reduced cellular tension doesn’t lead to altered vinculin binding as has been observed for other structural components (e.g. zyxin [155]).
Functions
Vinculin N-terminal, when bound to talin, partially opens up and aids integrin clustering for FA growth, possibly by retaining the activated state of integrins [156, 157]. The C-terminal forms a mechanosensitive link between adhesion receptors and the actin cytoskeleton to help recruit other components of the actin linking module (e.g. α-actinin, paxillin) and influence the mechanical strength of the cell [143, 157, 158]. Additionally, vinculin possesses actin filament capping activity [159]. This needs an complete opening of vinculin structure allowing C-terminal of the tail to compete with formin mDia1 for actin barbed ends.Vinculin also contributes to stability of focal adhesion under high forces by regulating contractile stress generation [143], thereby influencing the cell migration speed [160]. Cells deficient in vinculin cannot form lamellipodia, assemble stress fibers, or spread efficiently over a substrate [157].
Zyxin[Edit]
Zyxin is enriched along actin filaments, stress fiber bundles, and at cell-cell or cell-matrix adhesion sites [161, 162].  Figure 14. Zyxin: This schematic diagram illustrates the molecular organization of zyxin and provides examples for how zyxin is represented in figures throughout this resource.Zyxin is specifically found in more mature adhesions [163] and its absence in early adhesions is commonly used to distinguish the ‘age’ of an adhesion [164]. The main function of zyxin is to form a bridge between the adhesion components at the cell membrane and the internal cytoskeleton (reviewed in [165]). Zyxin is vital for coordinating matrix-dependent cues with actin dynamics; for example, within stress fibers and focal adhesions (FAs), zyxin acts as a mechanosensor that binds to areas where forces are applied [9, 10, 11]. Not only is zyxin binding proportional to the mechanical force (e.g. decreased traction reduces zyxin-binding) but its stability at adhesion sites is tension-dependent [155, 166].
Figure 14. Zyxin: This schematic diagram illustrates the molecular organization of zyxin and provides examples for how zyxin is represented in figures throughout this resource.Zyxin is specifically found in more mature adhesions [163] and its absence in early adhesions is commonly used to distinguish the ‘age’ of an adhesion [164]. The main function of zyxin is to form a bridge between the adhesion components at the cell membrane and the internal cytoskeleton (reviewed in [165]). Zyxin is vital for coordinating matrix-dependent cues with actin dynamics; for example, within stress fibers and focal adhesions (FAs), zyxin acts as a mechanosensor that binds to areas where forces are applied [9, 10, 11]. Not only is zyxin binding proportional to the mechanical force (e.g. decreased traction reduces zyxin-binding) but its stability at adhesion sites is tension-dependent [155, 166].
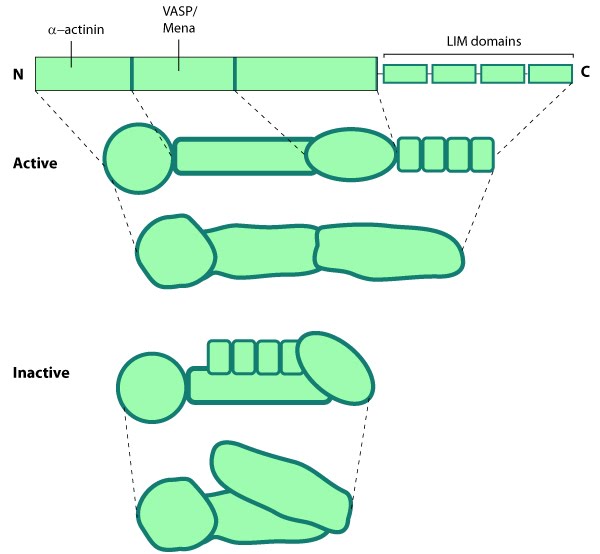 Figure 14. Zyxin: This schematic diagram illustrates the molecular organization of zyxin and provides examples for how zyxin is represented in figures throughout this resource.
Figure 14. Zyxin: This schematic diagram illustrates the molecular organization of zyxin and provides examples for how zyxin is represented in figures throughout this resource.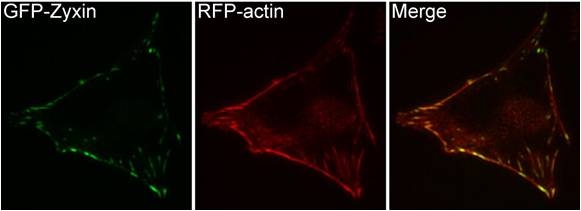 Figure 15. Zyxin localization: A NIH3T3 cell, plated on a collagen coated glass slide and transfected with RFP-actin and GFP-zyxin. The basal surface of the cell was imaged using TIRF microscopy, on an Olympus IX81 Inverted microscope at 60x magnification. [Image captured by Machiyama Hiroaki, Mechanobiology Institute, Singapore]
Figure 15. Zyxin localization: A NIH3T3 cell, plated on a collagen coated glass slide and transfected with RFP-actin and GFP-zyxin. The basal surface of the cell was imaged using TIRF microscopy, on an Olympus IX81 Inverted microscope at 60x magnification. [Image captured by Machiyama Hiroaki, Mechanobiology Institute, Singapore]